Open Tools & Data

.jpg)
This LifeCanvas SmartBatch+ protocol is intended for whole mouse brain tissue clearing and incorporates electrophoretic delipidation. While this delipidation protocol is longer than other delipidation protocols we tested, it preserves the endogenous fluorescence of the tissue and clears the whole brain most effectively, demonstrating the morphology of fine cell structures deep in the brain.
This protocol discusses how to mount the sample holder into the microscope and the steps to acquire a full-brain volumetric image using the custom ExA-SPIM software.
The Genetic Tools Atlas (GTA) is a searchable web tool representing information and data on enhancer-adeno-associated viruses (enhancer AAVs) and mouse transgenes developed and tested at the Allen Institute for Brain Science. The GTA offers a large genetic toolkit for selective gene expression in brain cell types of interest.
Code Ocean capsule for sharing analysis of thalamus MERFISH data, primarily in jupyter notebook form.
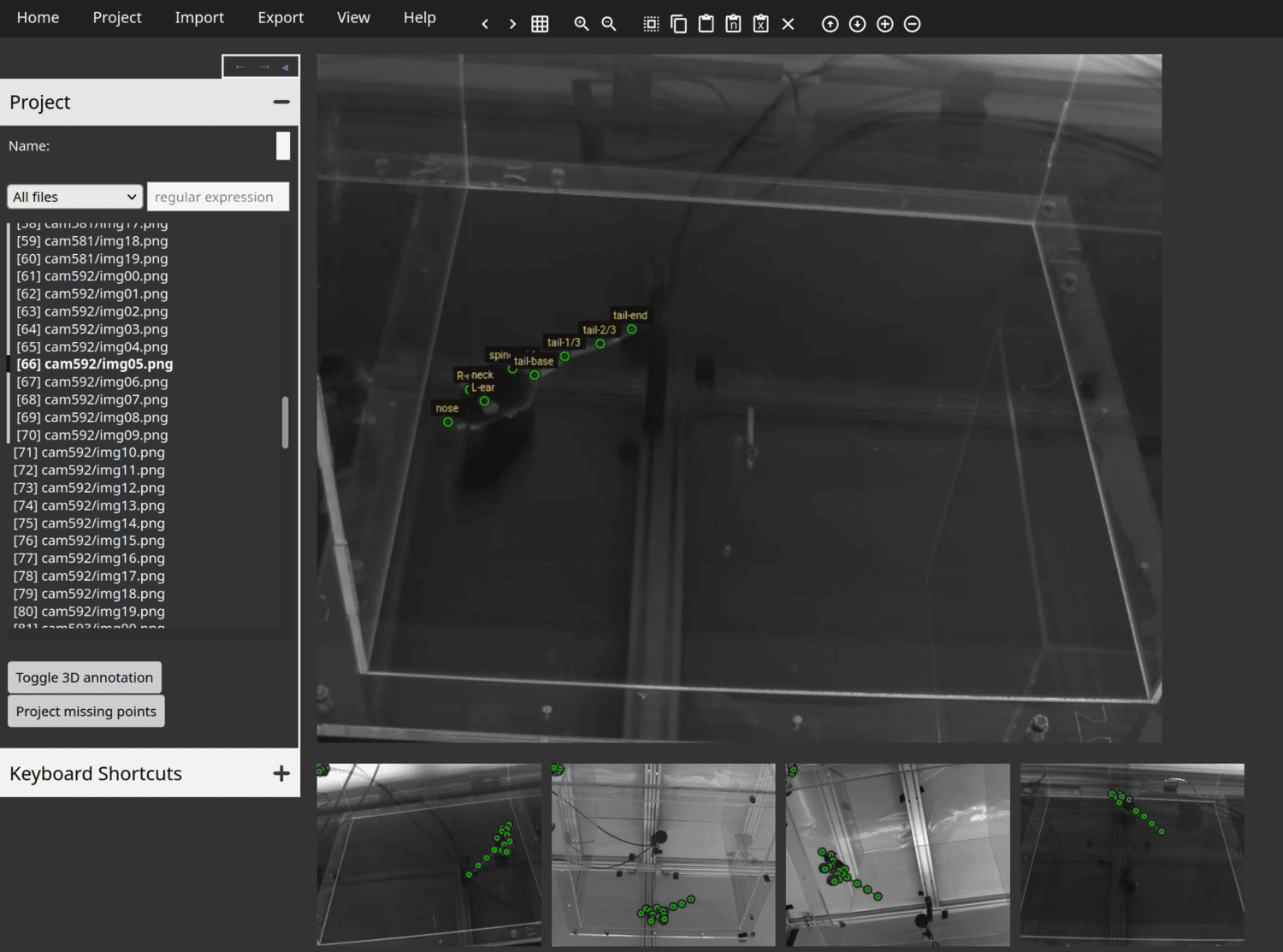
Anivia is a new web tool intended specifically for annotation of animal keypoints from images in both 2D and 3D.
A Harp core implementation on the RP2040
Several Harp devices:
On this repository you can find documentation of the Harp protocol, what is a Harp device and the Harp Timestamp Bus. The Harp technology was originally thought at Champalimaud Foundation to suppress the lack of efficient binary protocols for scientific hardware communication.
A low-level interface to data collected with the Harp binary protocol.

A library for managing data contracts and quality control in behavioral datasets.
A library for defining and maintaining behavior tasks.

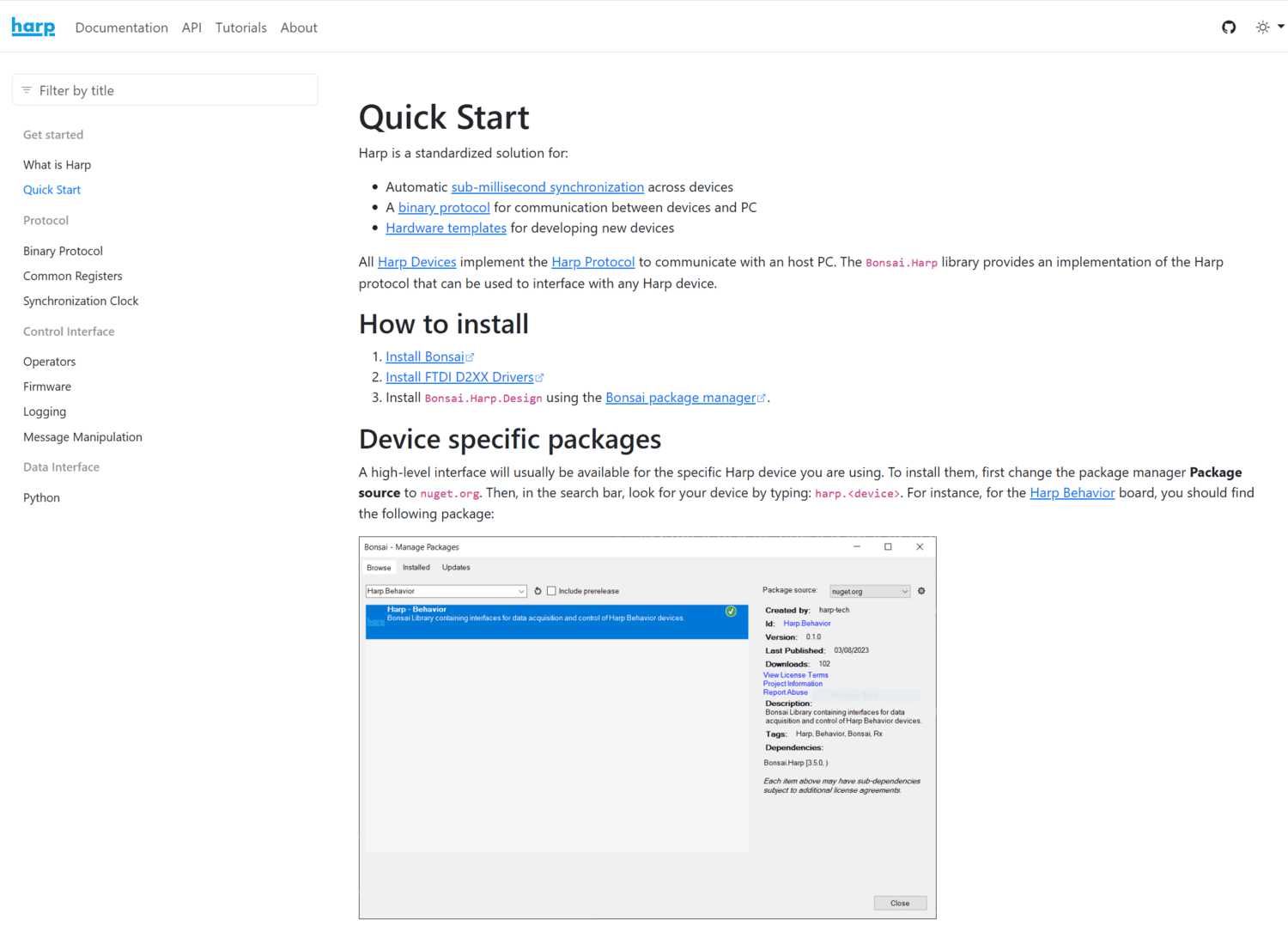
Harp is a standardized solution for:
All Harp Devices implement the Harp Protocol to communicate with an host PC. The Bonsai.Harp library provides an implementation of the Harp protocol that can be used to interface with any Harp device.
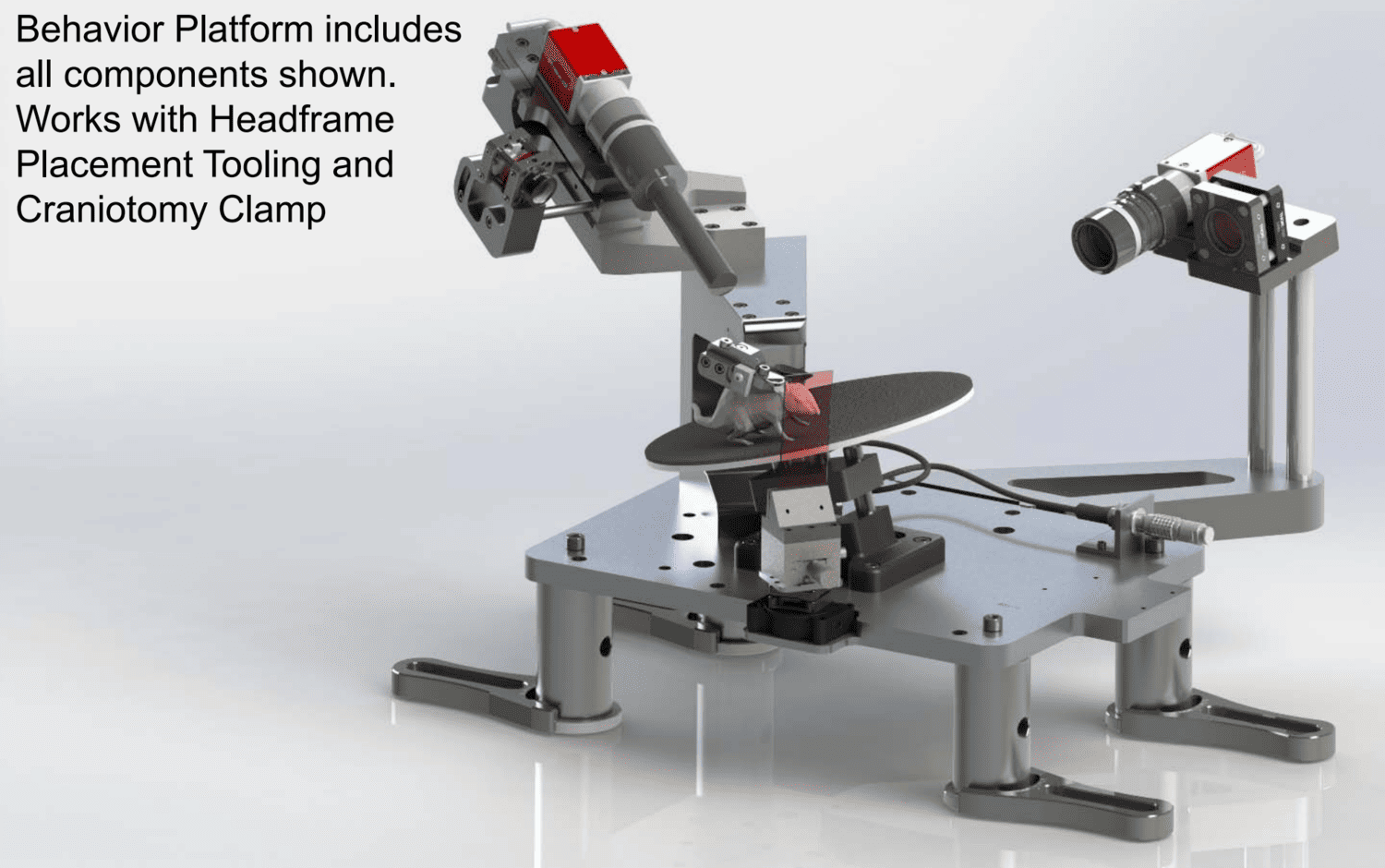
Included are the design specifications for a behavior platform that allow a headfixed mouse to behave freely on an adjustable inclined rotating disc, while keeping the right eye free of obstruction. The design interfaces to the Allen Institute Headframe assembly and includes the clamping mechanism required to mount the mouse.
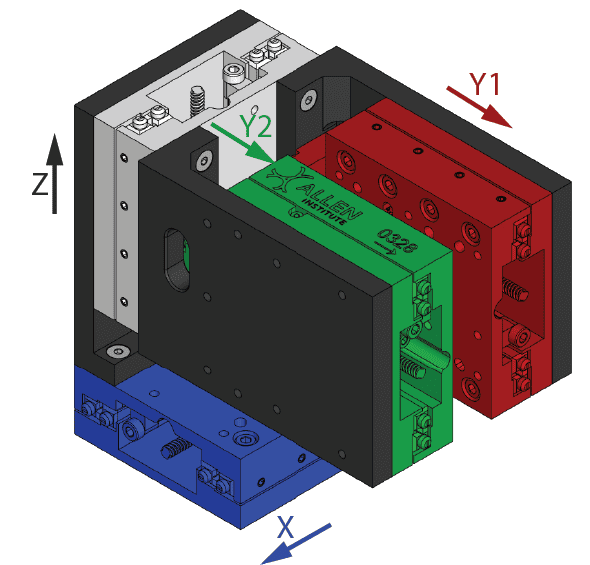
Software that controls the manipulator. The manipulator is composed of two main components:
A repository for calibration and configuration of the harp olfactometer device
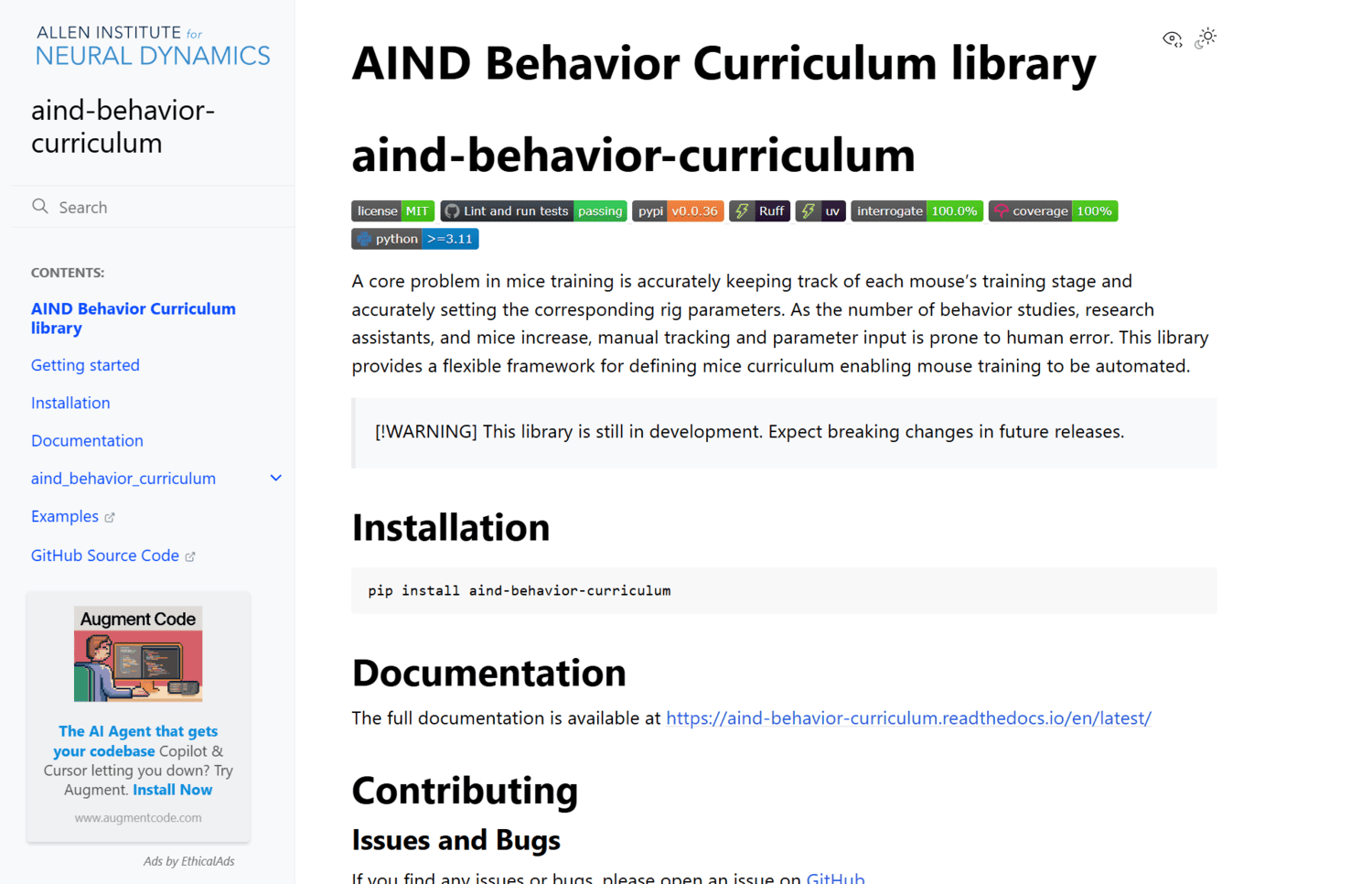
A core problem in mice training is accurately keeping track of each mouse’s training stage and accurately setting the corresponding rig parameters. As the number of behavior studies, research assistants, and mice increase, manual tracking and parameter input is prone to human error. This library provides a flexible framework for defining mice curriculum enabling mouse training to be automated.
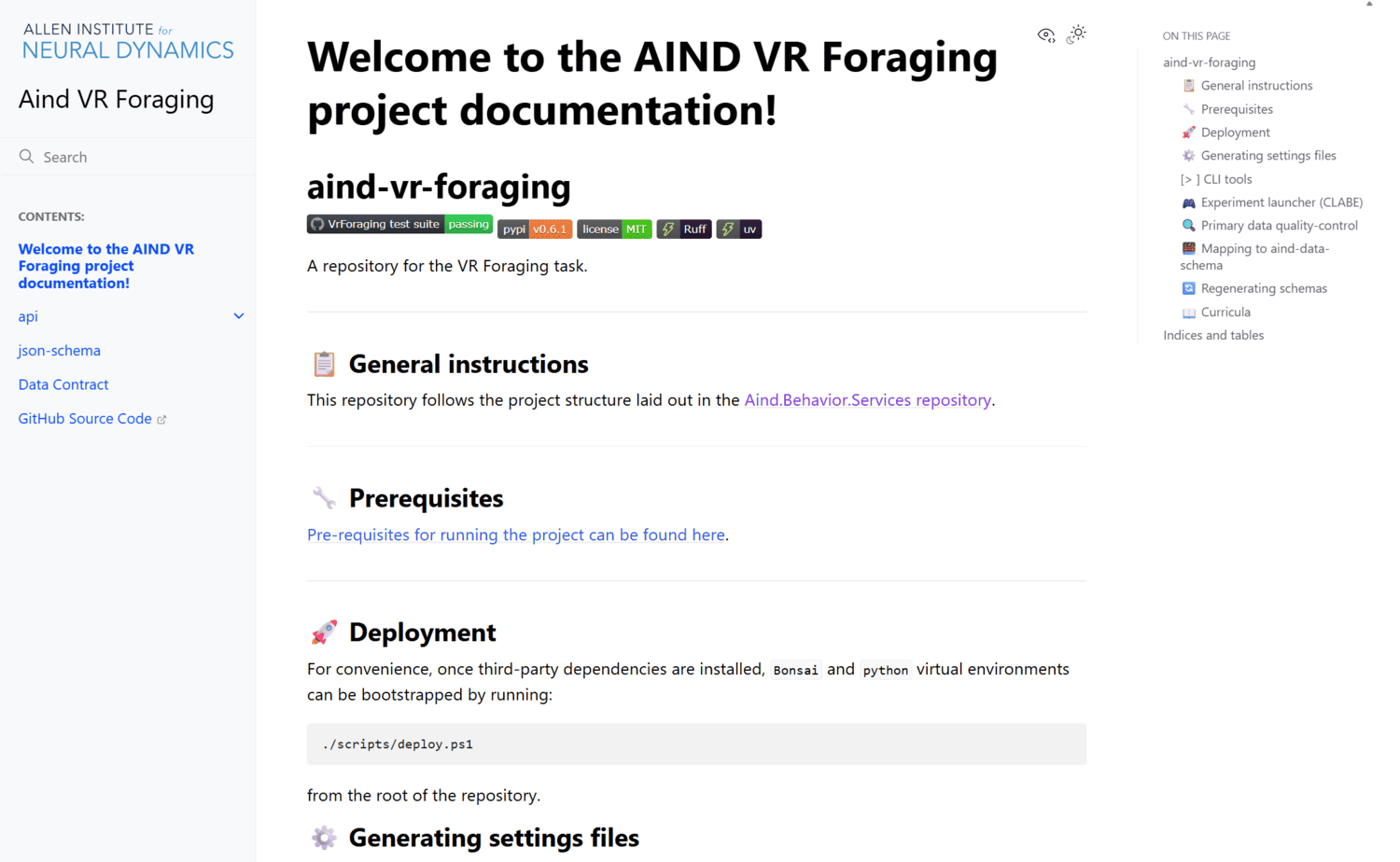
A repository for the VR Foraging task.
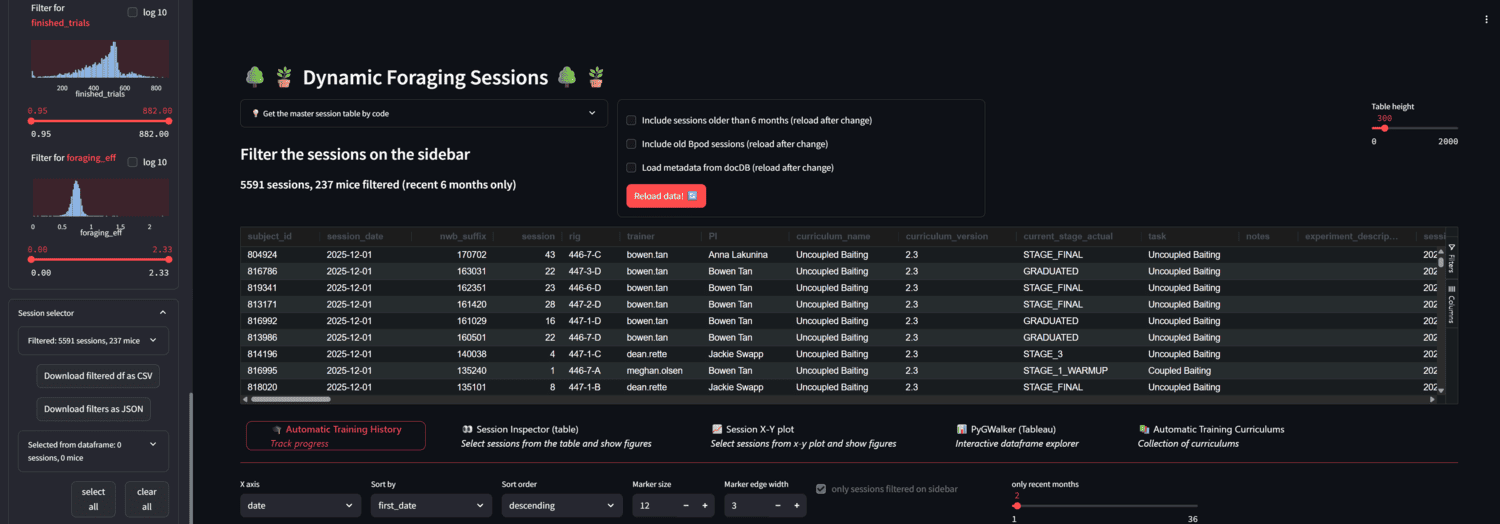
A streamlit app for browsing foraging behavior sessions in AIND.
A pubic collection accompanying "Efficient and reproducible pipelines for spike sorting large-scale electrophysiology data."

We present an end-to-end spike sorting pipeline that leverages parallelization to scale to large datasets.

This modified LifeCanvas embedding protocol is intended to prepare cleared mouse brains for imaging on the SmartSPIM. It details mixing and degassing the agarose solution and the technical process of embedding a brain in the agarose solution.

This protocol describes the experimental setup and procedure for behavioral training of mice in a “dynamic foraging” task, wherein a thirsty mouse is trained to make decisions in a dynamic environment.
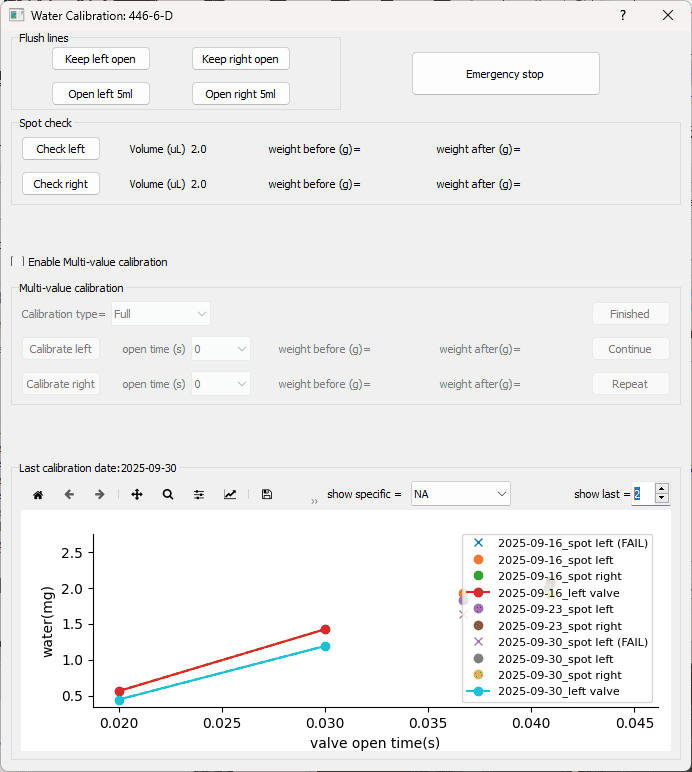
This protocol outlines the parameters, steps, and frequency of water calibration and spot checks to ensure that reward volumes remain consistent over time.
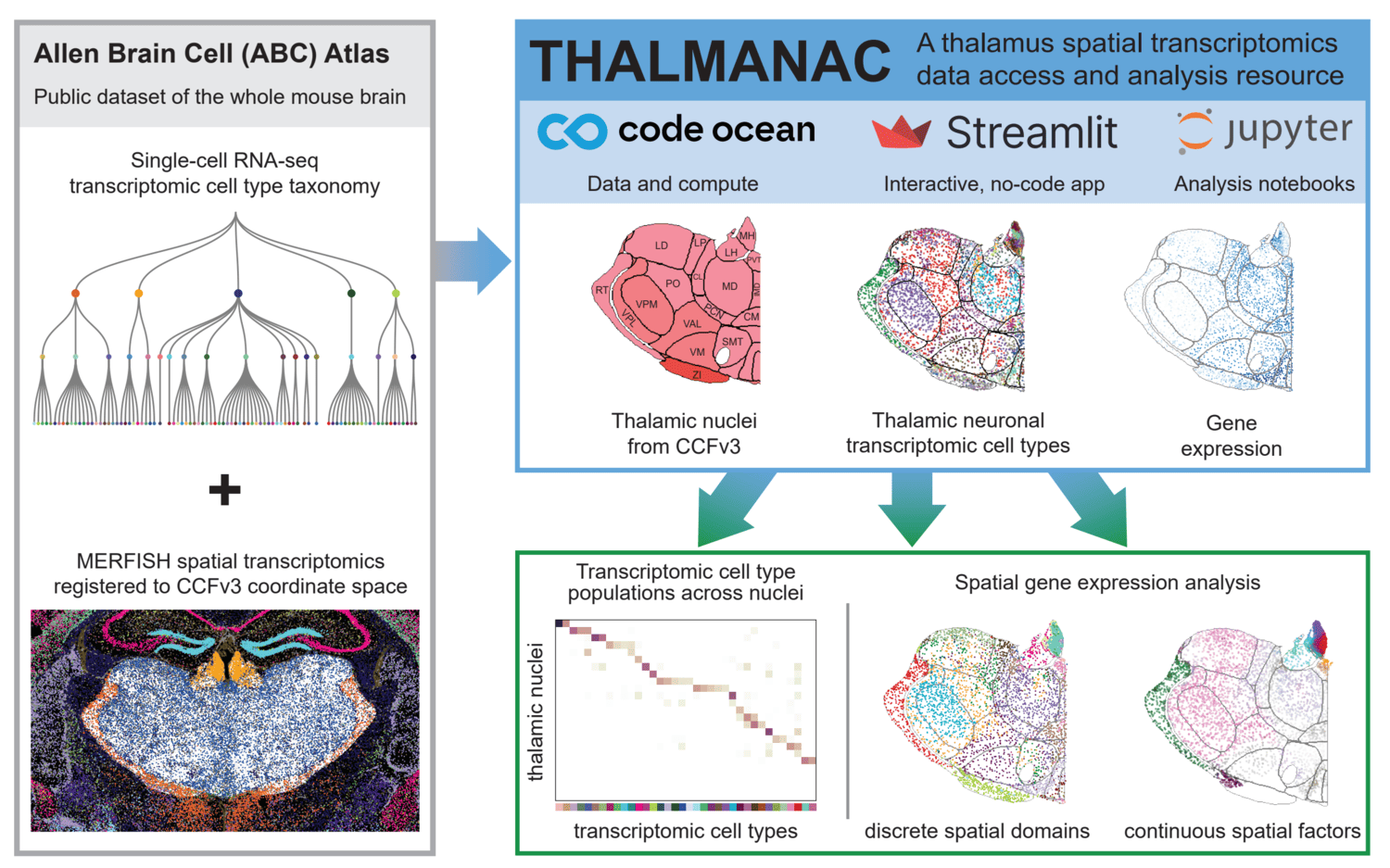
We present the THALMANAC, (THALamus MERFISH ANalysis and ACcess) a Findable, Accessible, Interoperable, Reusable and Reproducible (FAIRR) resource for exploring and analyzing single-cell transcriptomic variation in the thalamus. The THALMANAC provides streamlined access to thalamic gene expression data registered to the common coordinate framework and tools for quantitative analysis and visualization of these data, all encapsulated in a reproducible, cloud computing platform.

A Behavior platform designed for VR Foraging tasks, supporting multi-modal sensory inputs and integration with the Allen Institute headframe system.

This protocol describes the delivery of a neuronal tracer using the iontophoretic method. The surgery uses a stereotaxic system to target specific brain coordinates in the mouse.

This protocol describes the delivery of a neuronal tracer using the Nanoject II. The surgery uses a stereotaxic system to target specific brain coordinates in the mouse.

This protocol describes the surgical procedure, instrumentation, and reagents necessary for the installation of a stabilizing headframe and cranial window over the cortex of an adult mouse. This protocol should be utilized for neural imaging and experimentation regarding the mouse cortex.

A behavior platform designed for Dynamic Foraging tasks, supporting multi-modal sensory inputs and integration with the Allen Institute headframe system.
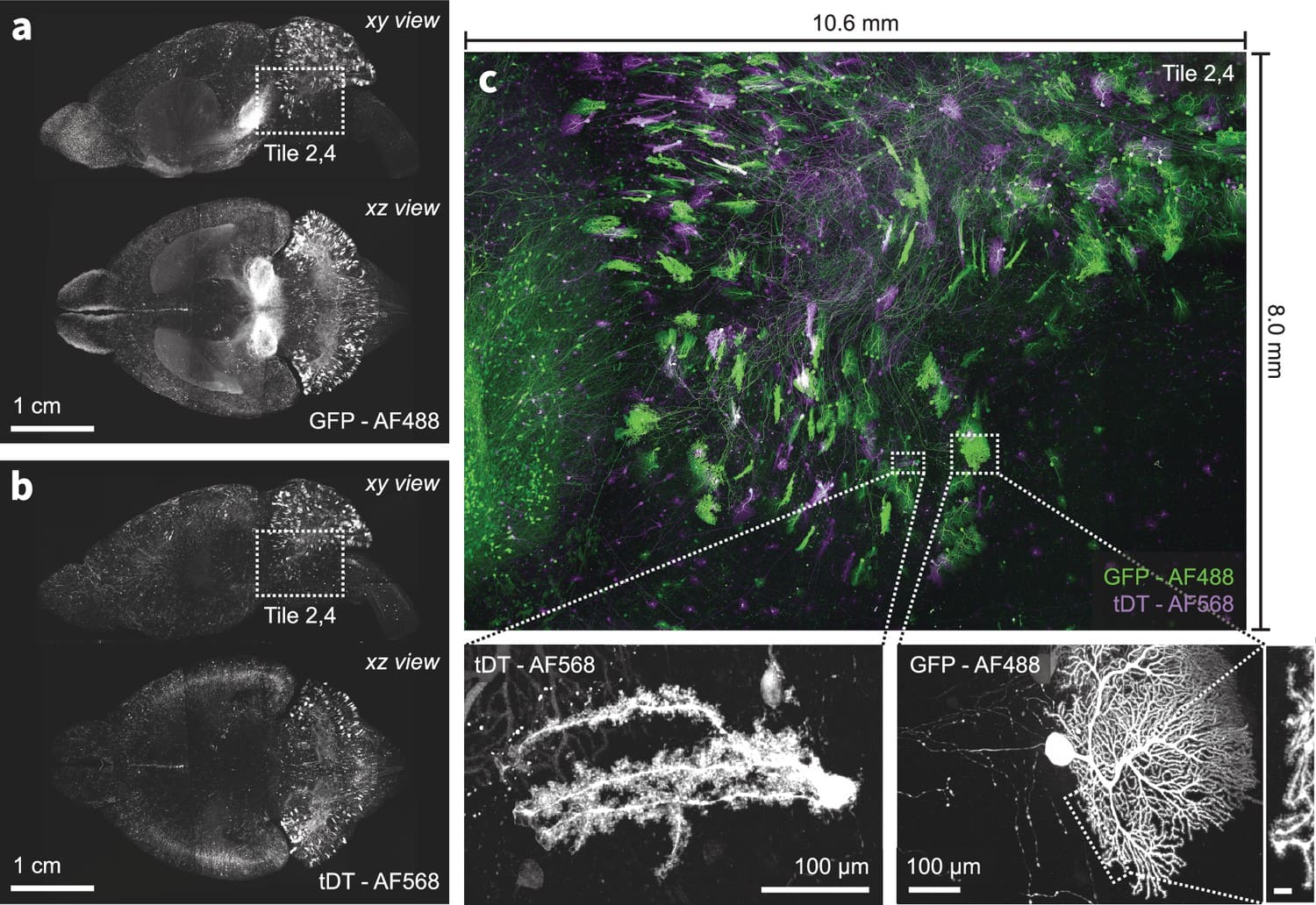
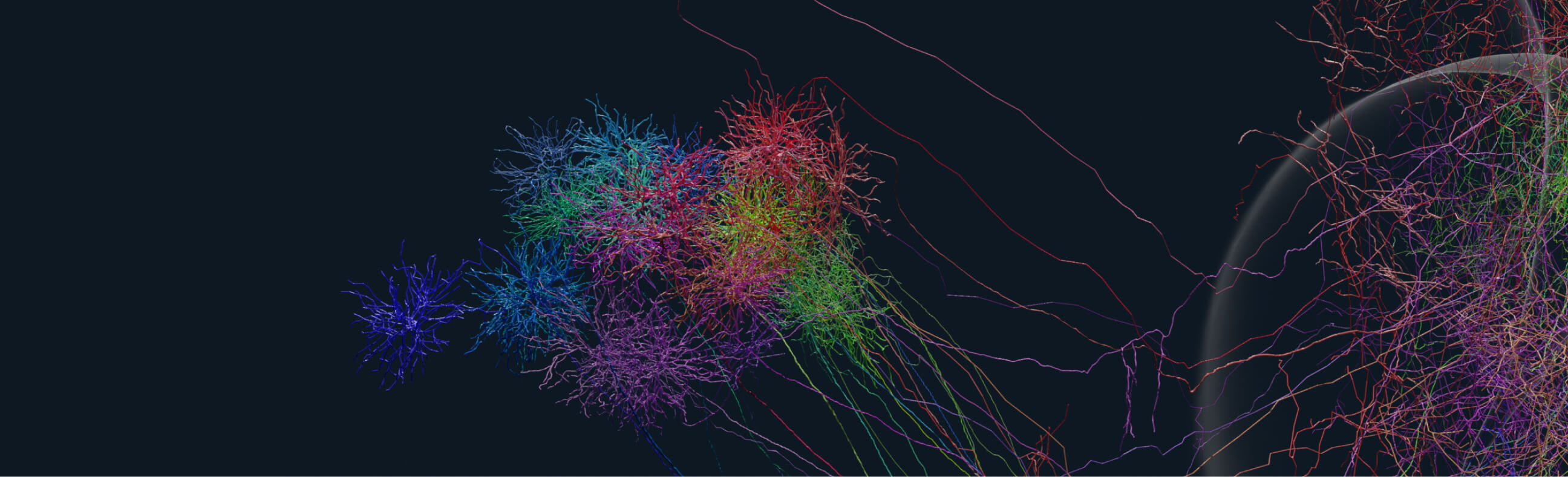
We present a new expansion-assisted selective plane illumination microscope, which, combined with new tissue clearing and expansion methods, the microscope allows imaging centimeter-scale samples with 250×250×750 nm optical resolution (4× expansion), including entire mouse brains, with high contrast and without sectioning.

BARseq is a high throughput in situ transcriptomics method utilizing designed barcodes incorporated in the amplification process for multiplexed gene detection.

Predictive processing theorizes that the brain continuously predicts sensory inputs, refining neuronal responses by minimizing prediction errors. Our collaborative experiments test these mechanisms using mismatch stimuli in mice and primates. The resulting datasets are shared via the OpenScope program and from collaborating laboratories, fostering community analysis and iterative refinement of predictive processing models.
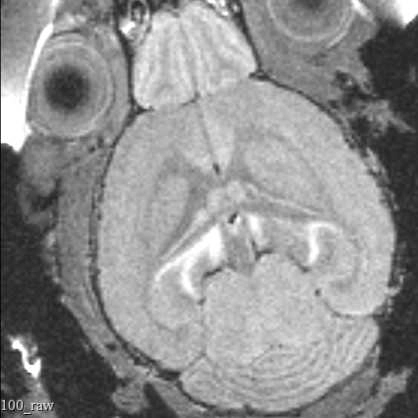
This protocol describes the process of obtaining pre-insertion MRI images using the University of Washington 14T vertical bore Bruker MRI. These structural scans allow for precisely targeting acute electrophysiology recordings in head-fixed mice beyond what is possible using skull landmarks alone.

The OpenScope program publishes on an annual basis a detailed public report that summarizes all of our activities within the year. These reports highlight all developments of the year: new selected projects, completed projects, publications and technical developments of the platform.

Frame-projected independent fiber photometry (FIP) is a method to measure fluorescent sensor signals through optical fibers implanted in living animals, using a camera to record video of the fiber faces. To precisely control timings of excitation and camera frame acquisition for FIP, and of other external stimulation apparatus (e.g. LEDs for photo-stimulation), we use a Teensy 4.1 microcontroller, which generates voltage pulses without relying on continuous communication with the operating system of the experimental computer for timing. This protocol is for the 3 excitation LED + 2 collection CMOS cameras design.
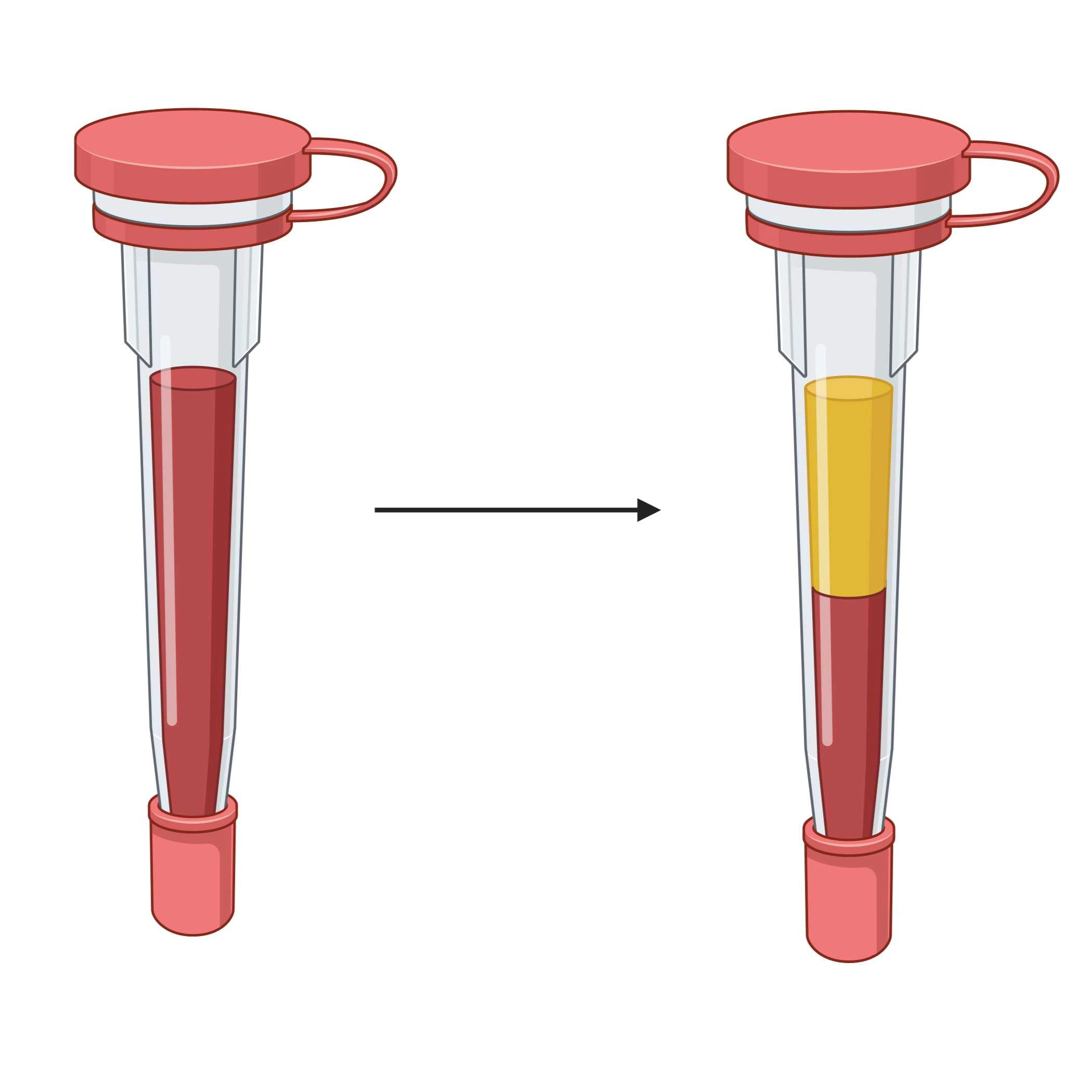
This protocol describes blood preparation steps required for analysis of cytokines using an Olink assay. This protocol is applied to samples collected during the protocol 'Temporal Assessment of Immune Response.'

This protocol describes Vascular Access Button (VAB) catheter blood collection intended for later assessment of immune responses across time to different IP injected substances in awake mice. This protocol is preceded by “Mouse Catheter Maintenance” protocol.

This protocol describes how to perform catheter maintenance to preserve mouse catheter patency after Vascular Access Button (VAB) implant.
This protocol describes the steps for preparing 20X stock and experimental solution of Lipopolysaccharide (LPS) intended for intraperitoneal injections in mice at a concentration of 0.25 mg/mL.

This protocol describes the surgical procedure, instrumentation, and reagents necessary for performing stereotactic injections and securing a headframe to an adult mouse brain for in-vivo recording procedures.
.jpg)
This is a step-by-step protocol to build a modified FIP (Frame-projected Independent Fiber Photometry) system. FIP was first implemented and reported by Kim et al. (2016). Their protocol is available here.
We have modified the previous design so that it can be built primarily with Thorlabs products (some products from other manufacturers are still required), which are off-the-shelf and easy to obtain with a reasonable lead time. The system described in this protocol is designed to record signals from 1) GFP-based sensors, 2) mApple-based sensors, and 3) isosbestic signals from up to 4-9 sites depending on fiber bundle selection (Fig. 1 and 2). When applied to measurements of other fluorescent protein-based sensors or spectrally shifted sensors, the selection of optical filters and/or excitation light sources should be modified accordingly. Also, the selection of the fiber patch-cable should be based on the number of simultaneously recorded sites and the diameter/NA of the fiber implants.

PREPRINT: Identifying the input-output operations of neurons requires measurements of synaptic transmission simultaneously at many of a neuron’s thousands of inputs in the intact brain. To facilitate this goal, we engineered and screened 3365 variants of the fluorescent protein glutamate indicator iGluSnFR3 in neuron culture, and selected variants in the mouse visual cortex. Two variants have high sensitivity, fast activation (< 2 ms) and deactivation times tailored for recording large populations of synapses (iGluSnFR4s, 153 ms) or rapid dynamics (iGluSnFR4f, 26 ms). By imaging action-potential evoked signals on axons and visually-evoked signals on dendritic spines, we show that iGluSnFR4s/4f primarily detect local synaptic glutamate with single-vesicle sensitivity. The indicators detect a wide range of naturalistic synaptic transmission, including in the vibrissal cortex layer 4 and in hippocampal CA1 dendrites. iGluSnFR4 increases the sensitivity and scale (4s) or speed (4f) of tracking information flow in neural networks in vivo.

XYZ sample collection platform with loop collection and integrated optical system

Microtome with slice thickness control and cutting actuation realized by high-precision linear stages

A reproducible system neuroscience notebook collection to facilitate data sharing and collaborative reuse with open science datasets
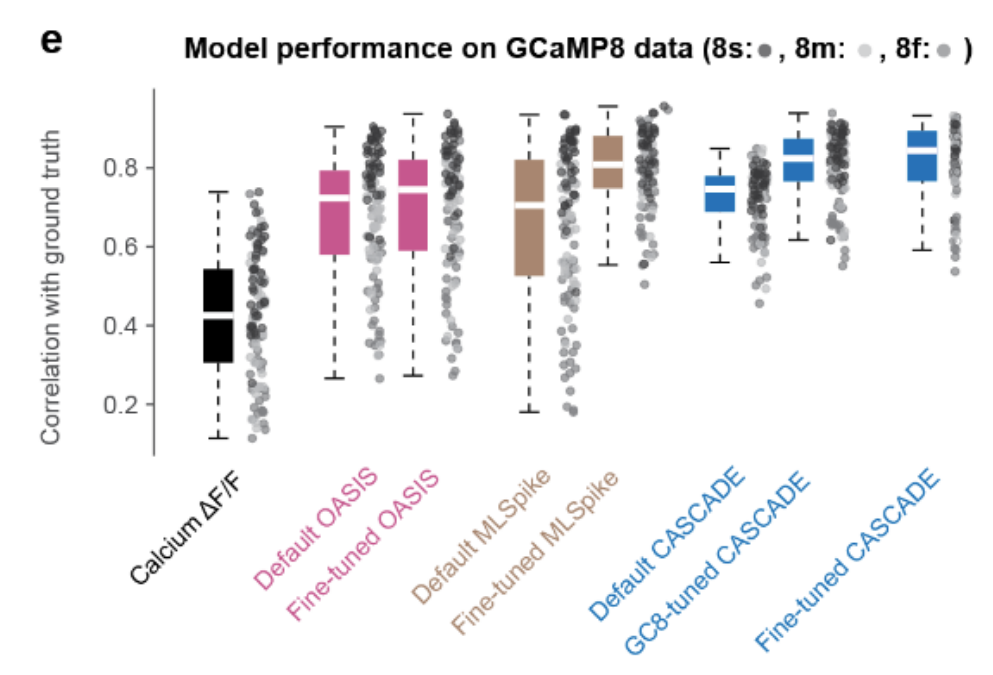
PREPRINT: Here, we evaluate the ability of the recently developed calcium indicator GCaMP8 to reveal neuronal spiking, and we investigate how existing spike inference methods (CASCADE, OASIS, MLSpike) should be adapted for optimal performance.
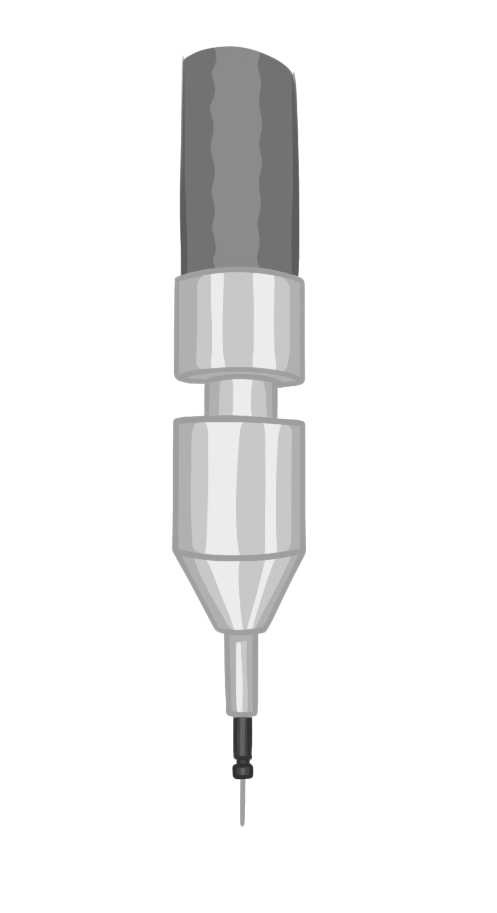
This protocol describes the surgical procedure, instrumentation, and reagents necessary for implanting optic fiber probes involving injection(s) and headpost into an adult mouse brain for in-vivo fiber photometry and/or optogenetic manipulation procedures.
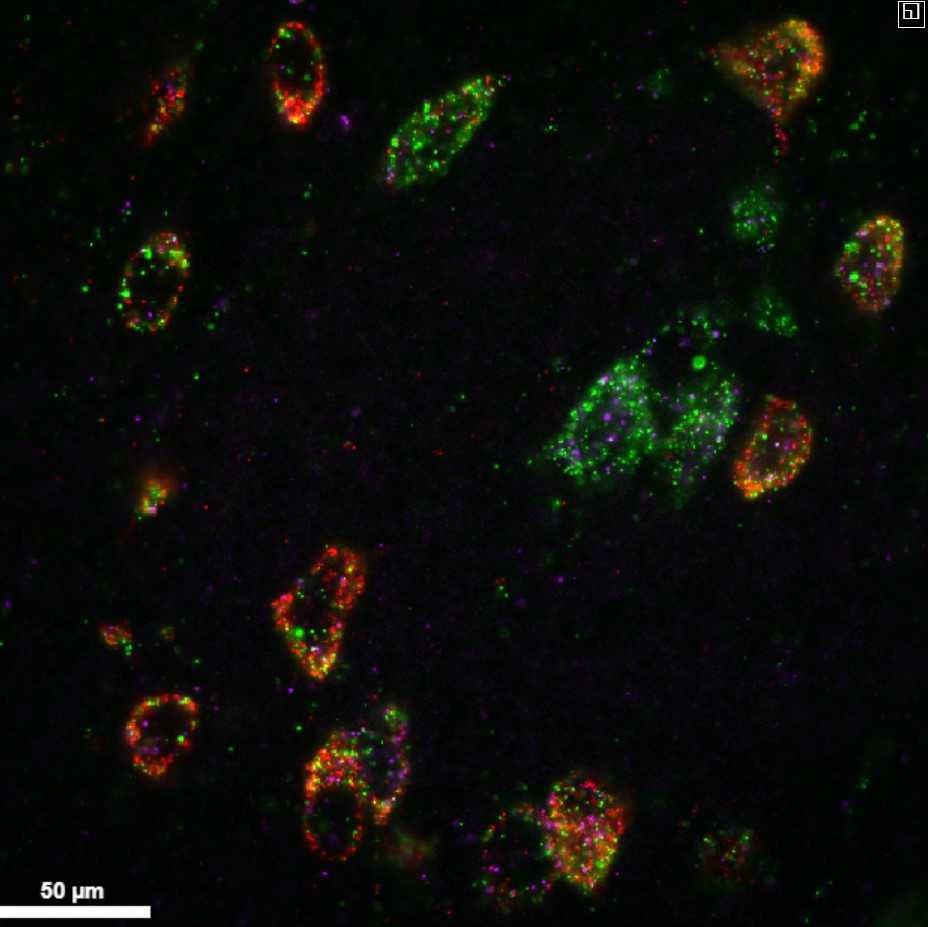
This protocol describes a method that utilizes expansion microscopy to perform multiple rounds of multiplexed fluorescent in situ hybridization (mFISH) on thick slices of brain tissue.
This protocol is modified from the Index Matching protocol found in the LifeCanvas Technologies Full Active Pipeline Protocol.

This protocol details index matching for delipidated whole mouse brain samples in ethyl cinnamate solution to prepare for lightsheet imaging.
PREPRINT: Neuropixels Opto probes represent an unprecedented tool for recording, identifying, and manipulating neuronal populations by combining high-resolution extracellular electrophysiology with optogenetics.
.jpg)
This protocol describes the typical workflow for the habituation of a mouse for awake behavioral experimentation that utilizes tubes to hold mice during head fixation.
.png)
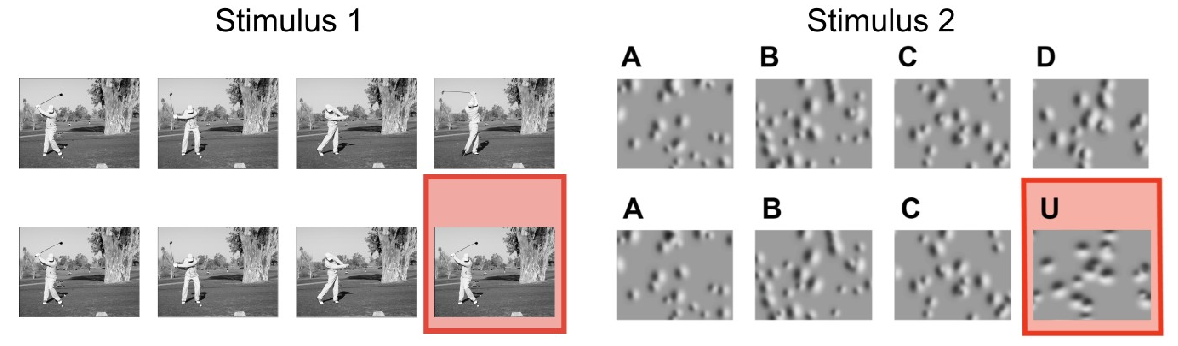
PREPRINT: Predictive coding (PC) hypothesizes that the brain computes internal models of predicted events and that unpredicted stimuli are signaled with prediction errors that feed forward. We tested this hypothesis using a visual oddball task. A repetitive sequence interrupted by a novel stimulus is a “local” oddball. “Global” oddballs defy predictions while repeating the local context, thereby dissociating genuine prediction errors from adaptation-related responses. We recorded neuronal spiking activity across the visual hierarchy in mice and monkeys viewing these oddballs. Local oddball responses largely followed PC: they were robust, emerged early in layers 2/3, and fed forward. Global oddball responses challenged PC: they were weak, absent in most visual areas, more robust in prefrontal cortex, emerged in non-granular layers, and did not involve inhibitory interneurons relaying predictive suppression. Contrary to PC, genuine predictive coding does not emerge early in sensory processing, and is instead exclusive to more cognitive, higher-order areas.

The visual cortex predicts incoming sensory stimuli through internal models that are updated following unexpected events. Cortical inhibitory neurons, particularly VIP interneurons, play a critical role in representing unexpected stimuli. Notably, this response is stimulus non-specific, raising the question of what information it conveys. Given their unique connectivity, we hypothesized that during unexpected stimuli, VIP neurons encode broad context signals, referred to here as task-independent information. To test this hypothesis, we analyzed the Allen Institute Visual Behavior dataset, in which mice viewed repeated familiar images and unexpected omissions of these images, while two-photon calcium imaging was performed from distinct cell types across primary and higher-order visual areas. Using dimensionality reduction methods, we found that, in contrast to image presentations, unexpected omissions trigger task-independent signaling in VIP and excitatory neurons. This signaling may facilitate the integration of contextual and sensory information, enabling updated predictions.
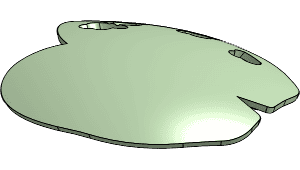
Single and dual hemisphere implant templates with supporting installation tools.
.png)
Headframe templates and designs for the Allen Institute headframe clamp system for intersession and interinstrument spatial registration.
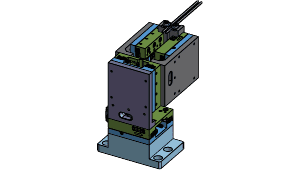
Compact, cost-effective, modular stimulus tool stage suitable for mouse behavior tasks.

New treadmill-style running wheel enables natural running posture, closed-loop variable effort running, headframe integration for a range of neurophysiology experiments, and measuring torque and speed of wheel.
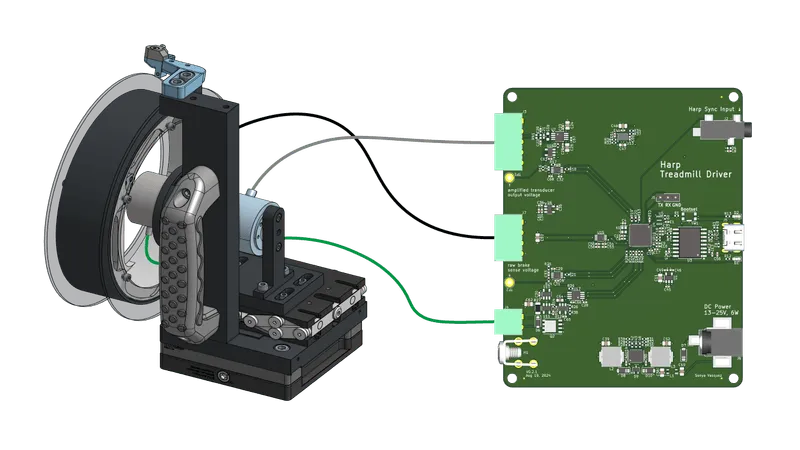
A Harp-compatible device for controlling an instrumented mouse treadmill.
Signal conditioning board for an implanted thermistor.
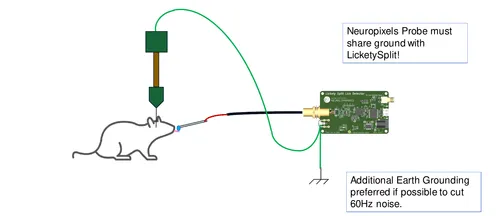
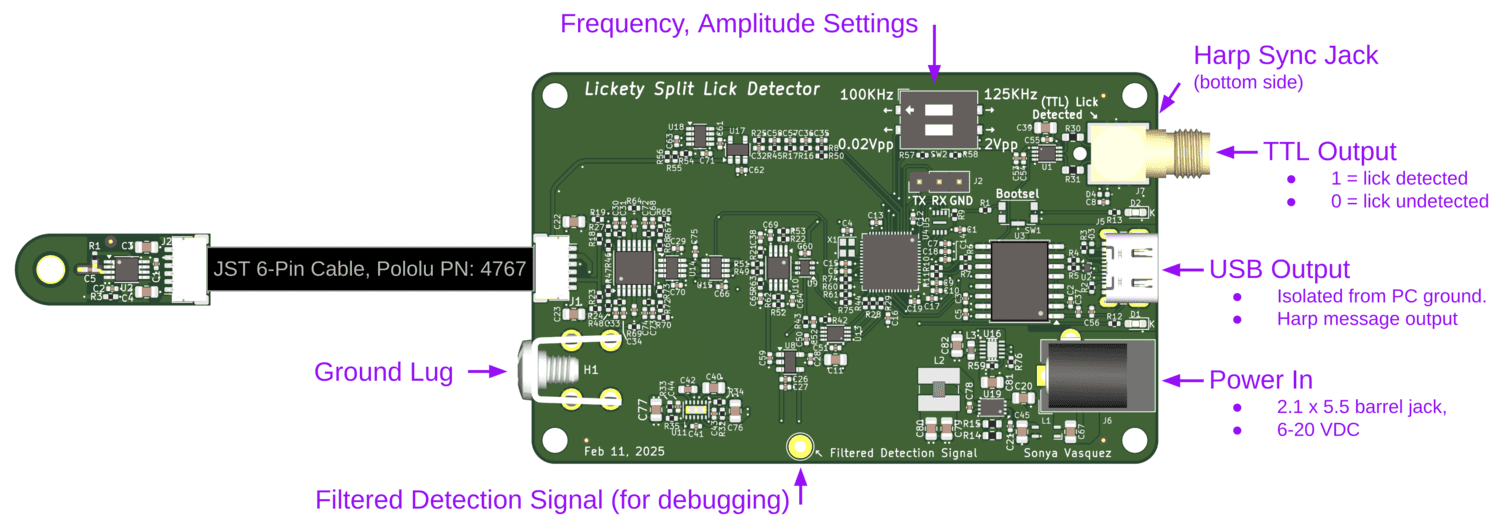
An ephys-compliant capacitive lick detector.
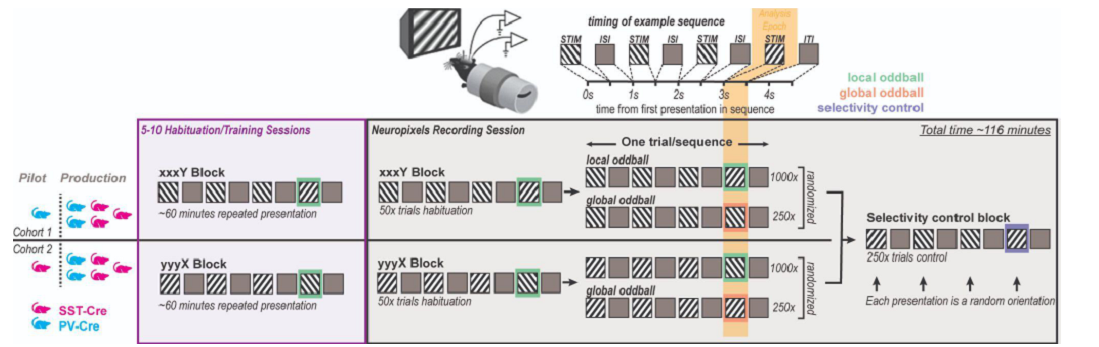
This dataset was collected for the Global Local Oddball project, as part of the OpenScope project.
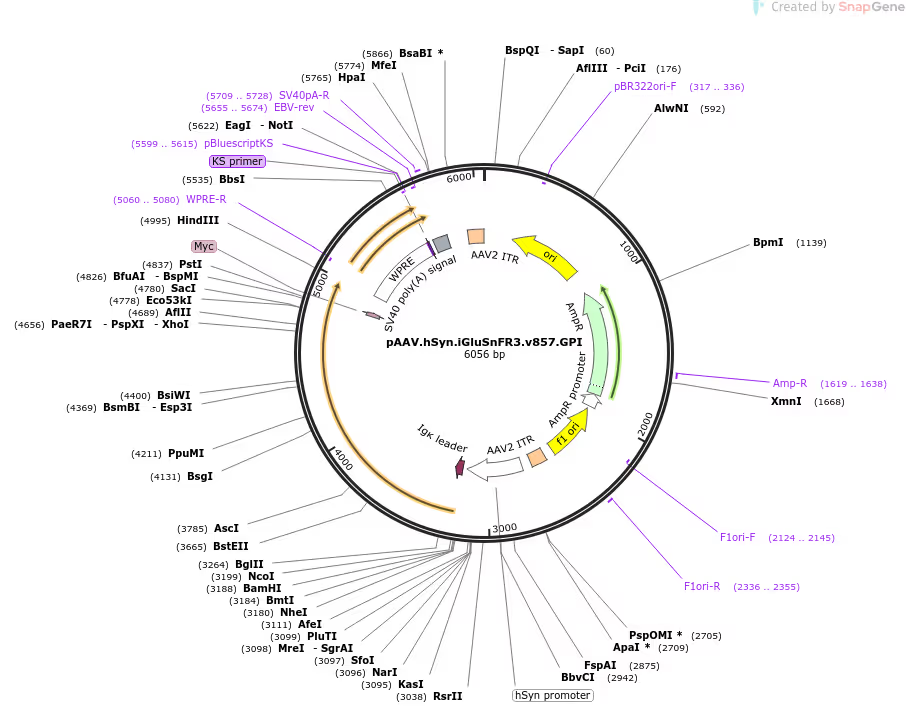
The Kaspar Podgorski Lab has deposited materials at Addgene for distribution to the research community. We are developing fluorescent indicators that report subthreshold inputs to directly read out intricate three-dimensional patterns of synaptic activity in behaving animals, so that we can infer how neurons transform inputs into outputs.
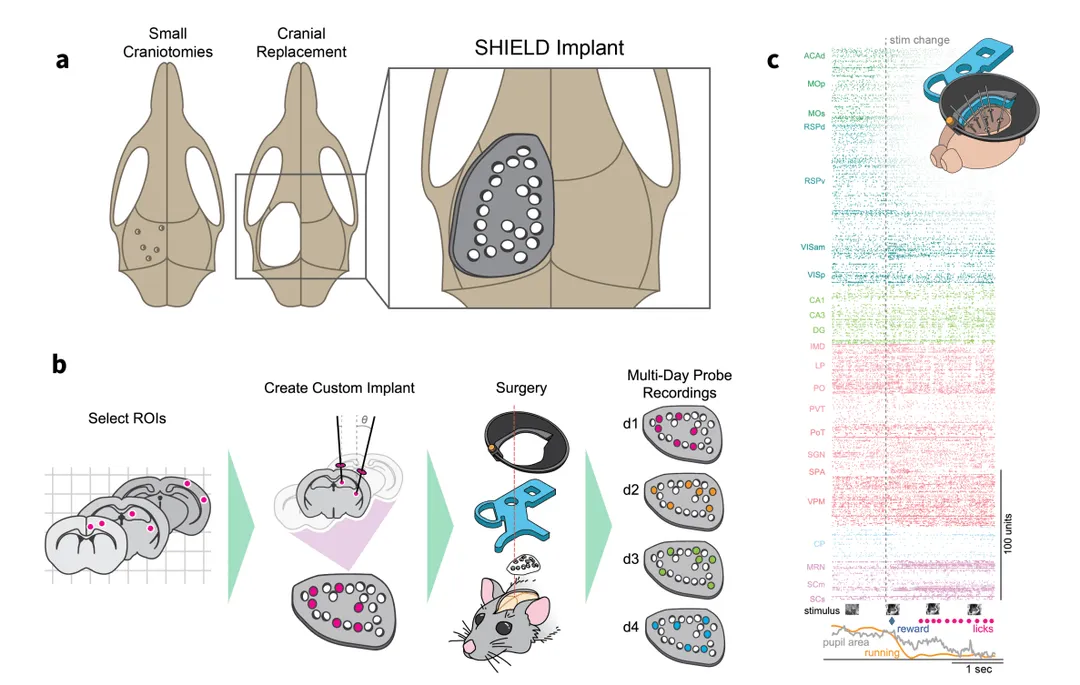
We describe a novel 3D-printed cranial-replacement implant (SHIELD) enabling electrophysiological recordings from distributed areas of the mouse brain, which allows us to measure spiking dynamics across many interacting brain regions.

This protocol describes the delipidation of a mouse brain specimen using a modified iDISCO protocol.
This protocol describes the pre-operative setup and post-operative take-down procedures utilized for rodent stereotaxic neurosurgical procedures.

This protocol describes the steps necessary to apply this silicone layer in advance of a cranial windowing procedure.
.avif)
This protocol describes a customized mouse habituation protocol for mice within experiments involving head fixation onto disks.
This protocol describes a semi-customized workflow to perform water restriction with mice designated for future experiments.
(1).avif)
This protocol describes the procedure for removing the SORTA-clear plug used in acute in vivo electrophysiology experiments in whole-hemisphere craniotomy (WHC) preparation mice with Neuropixels probes.
This protocol describes the procedure for making agarose used in acute in vivo Electrophysiology Experiments with Neuropixels probes.
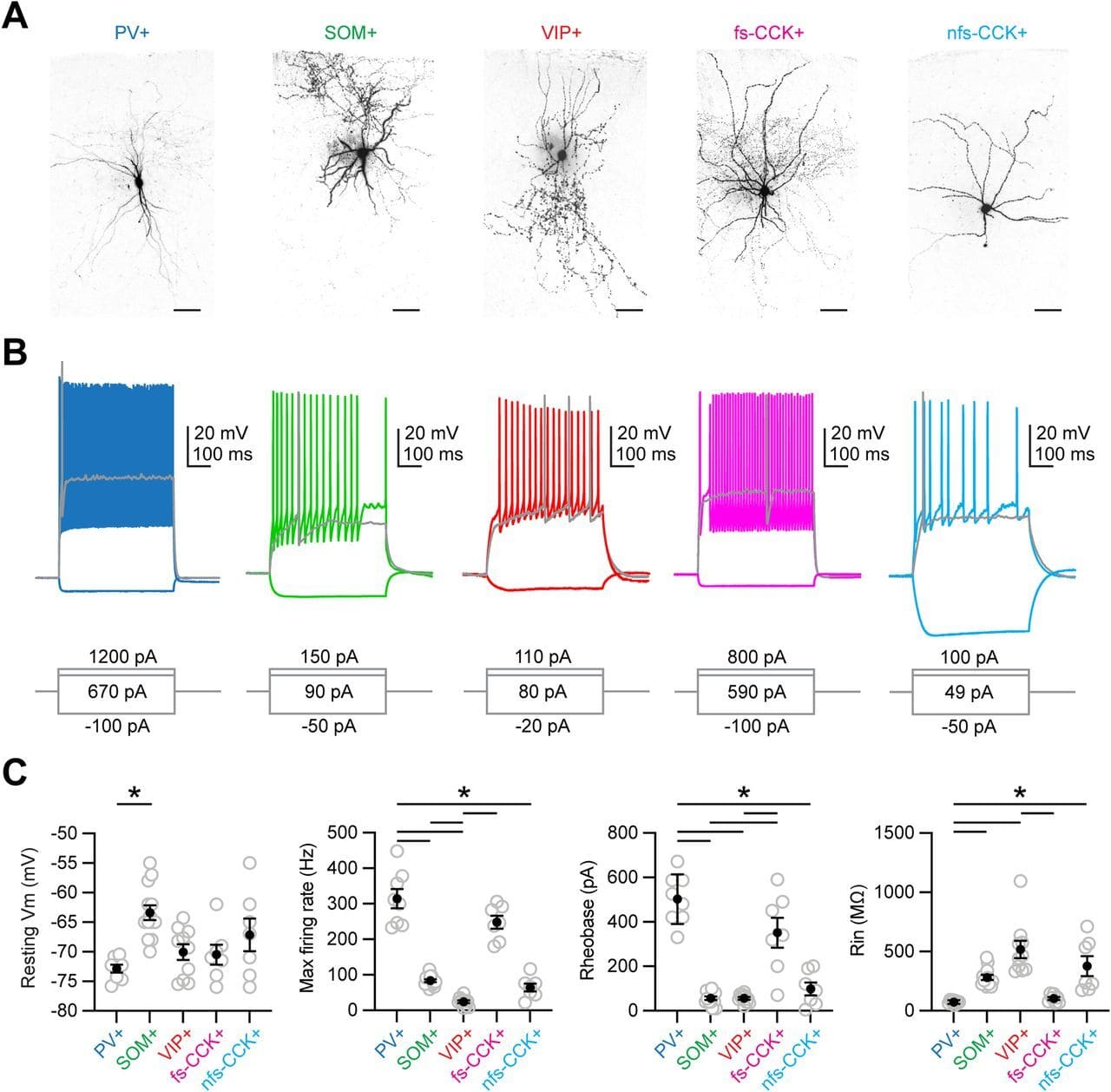
Here, we use anatomy, slice physiology, and optogenetics to examine how mediodorsal (MD) thalamus evokes feed-forward inhibition in Layer 3 of the prelimbic subregion of the mouse prefrontal cortex (PFC).
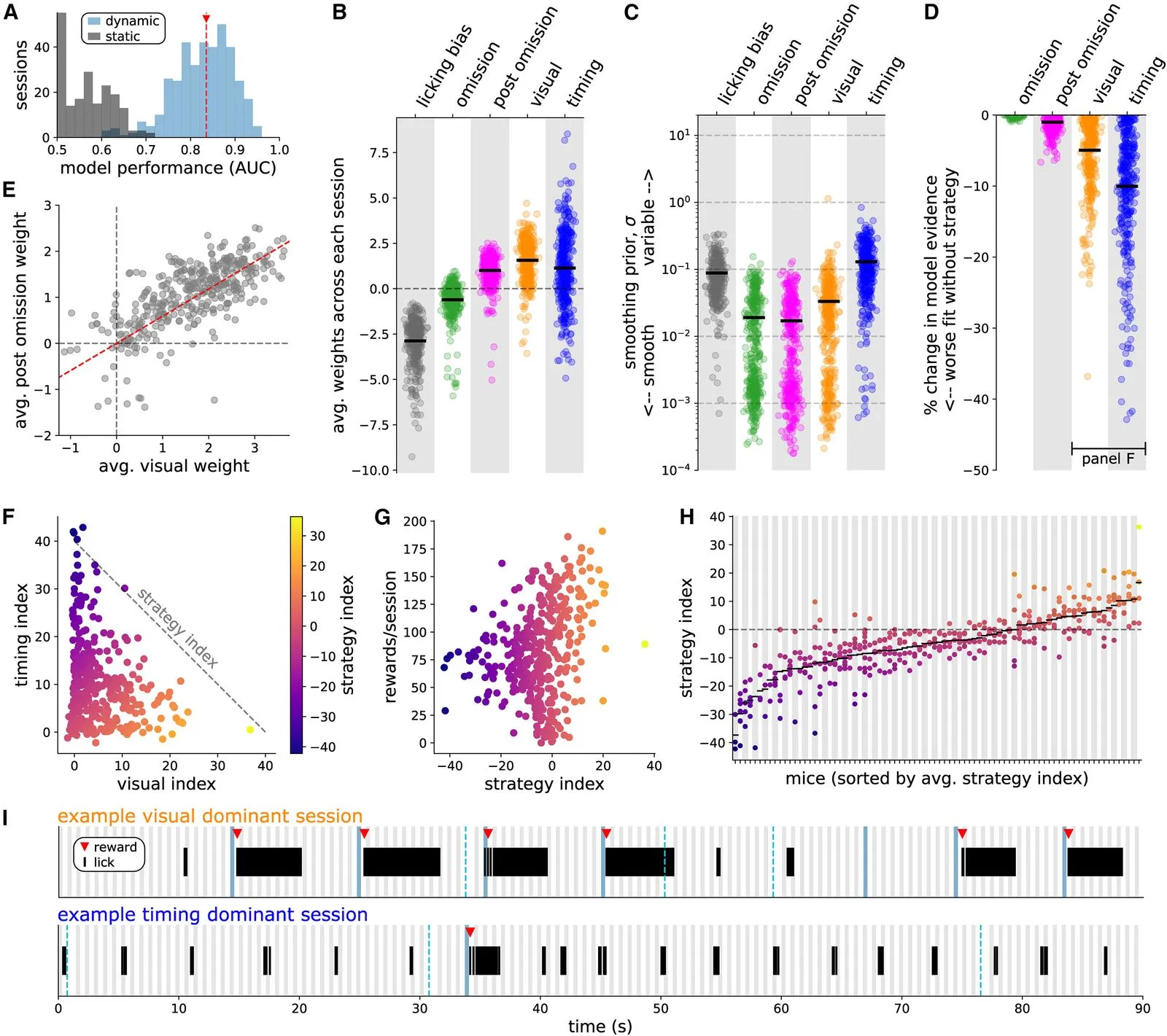
We investigate the question of how distinct animal reward strategies differentially engage brain circuits, focusing on the cortical Vip-Sst disinhibitory circuit between vasoactive intestinal peptide-postive (Vip) interneurons and somatostatin-positive (Sst) interneurons.
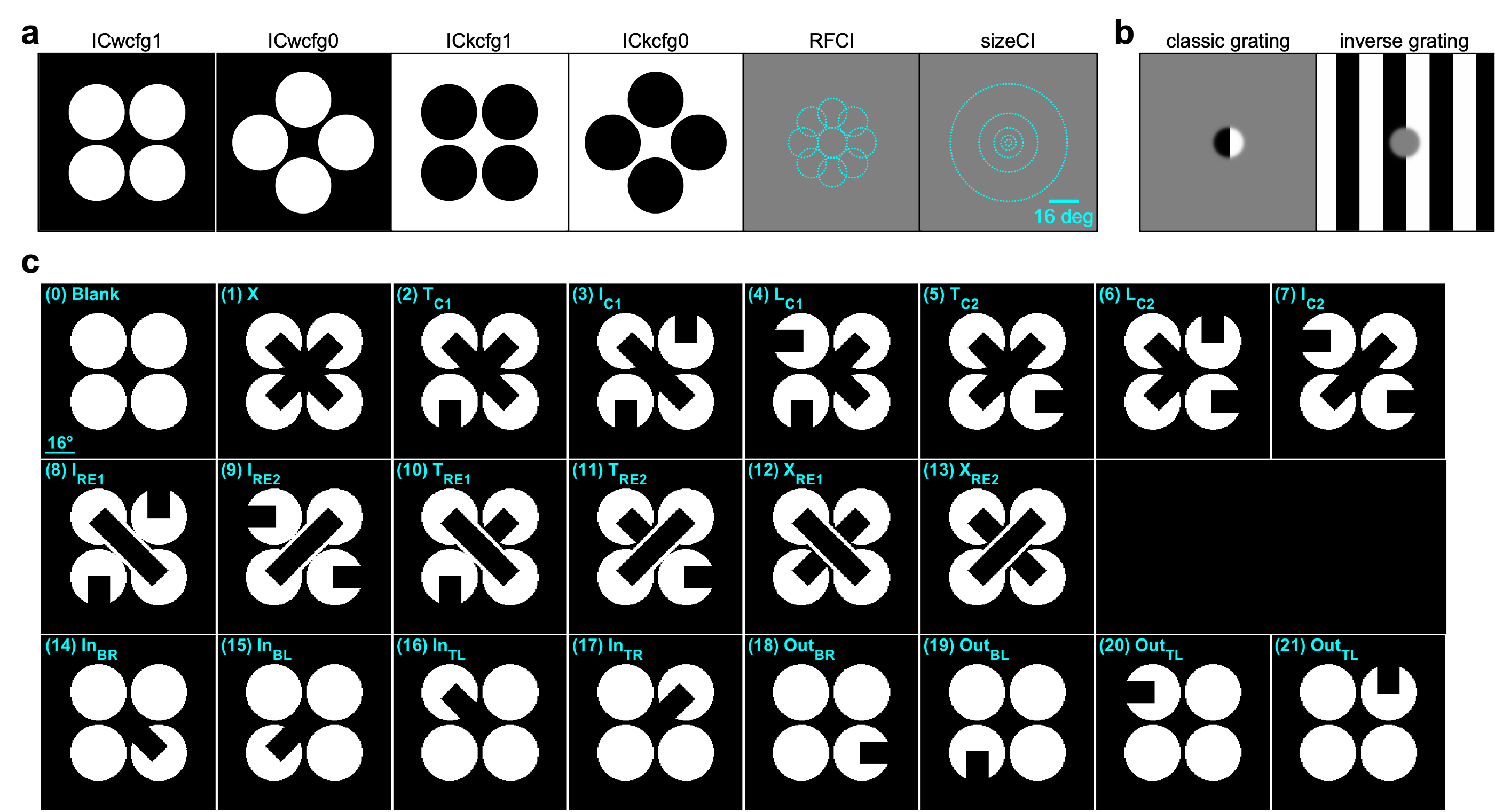
This dataset was collected for the Illusion project, as part of the OpenScope project.
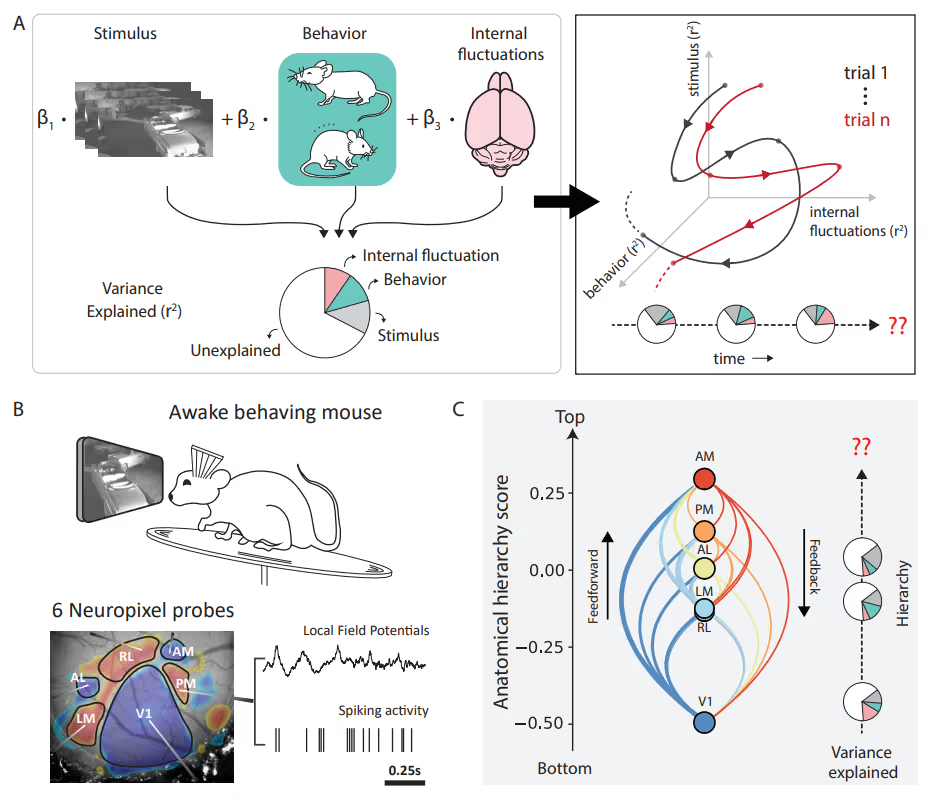
PREPRINT: To address the question of how factors' (such as brain states and behaviors) relative impact neuronal variability evolves over time, we designed an encoding model with latent states to partition visual cortical variability across three crucial categories of sources: internal brain dynamics, behavior, and external visual stimulus.
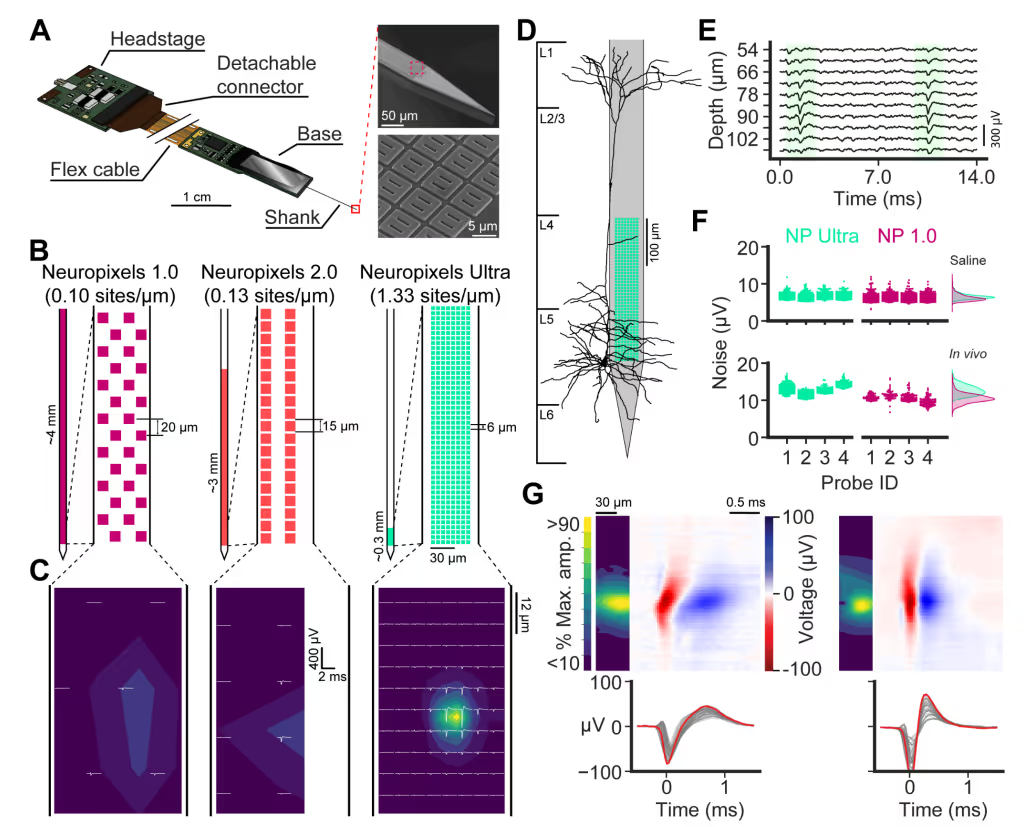
PREPRINT: We developed a silicon probe for extracellular electrophysiology which samples neuronal activity at ultra-high spatial density, allowing us to more accurately and sensitively measure neural activity.
This protocol describes the process of coating an implant with duragel in preparation for in vivo electrophysiology experiments.
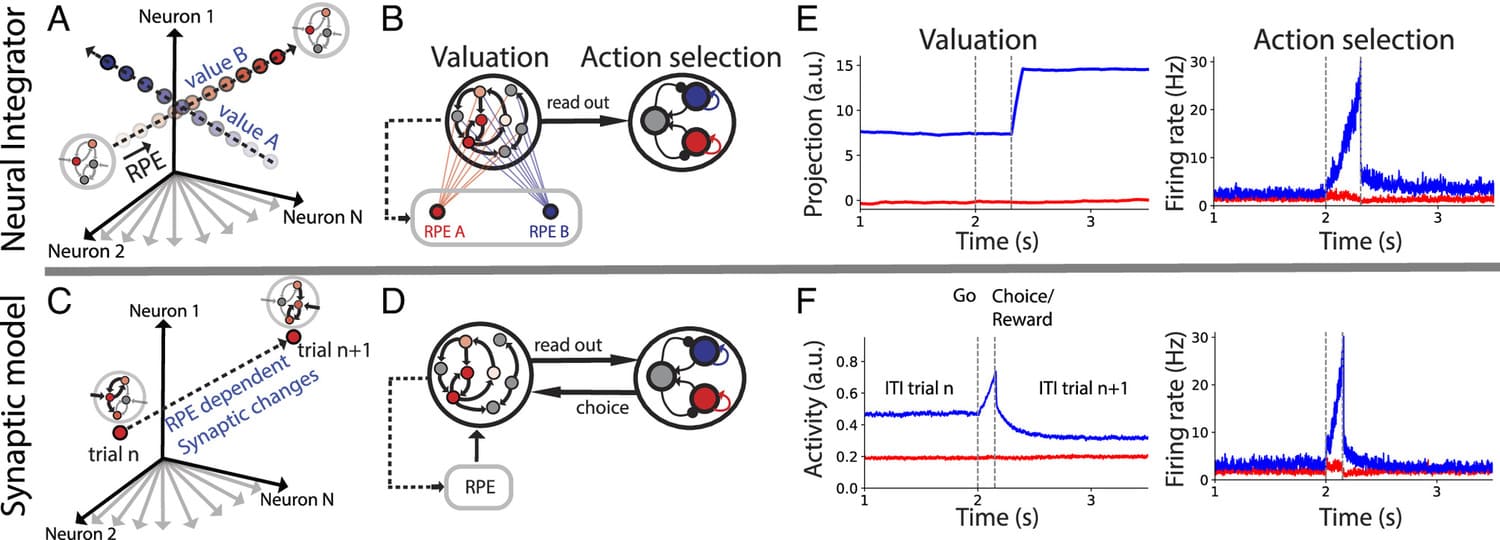
During foraging behavior, action values are persistently encoded in neural activity and updated depending on the history of choice outcomes. What is the neural mechanism for action value maintenance and updating? Here, we explore two contrasting network models: synaptic learning of action value versus neural integration.

This protocol details mounting thin sliced mouse brain tissue sections onto charged slides in a uniform orientation that does not create wrinkles or artifacts that might interfere with imaging, and then how to apply a coverslip.

The Immunohistochemistry (IHC) Staining for Mouse Brain Sections protocol details the blocking, primary, and secondary antibody staining of 50-100 micron mouse brain tissue slices fixed in 4% PFA.
This protocol details the steps for staining 4% PFA fixed mouse brain tissue sections with DAPI (4',6-diamidino-2-phenylindole), a fluorescent stain that can be used to label anatomic regions of interest in a mouse brain, allowing for imaging with a fluorescent microscope.

This is a protocol that details sectioning a mouse brain fixed in 4% PFA and 30% sucrose on a sliding microtome.

This protocol collection details the steps for a whole mouse brain specimen to be perfused, sliced, stained with antibodies, DAPI, or both, and made into slides.
A Bonsai workflow for lick-based foraging experiments, with a PyQt frontend for visualizing performance and modifying task parameters.

This dataset was collected for the Credit Assignment project, as part of the OpenScope project.
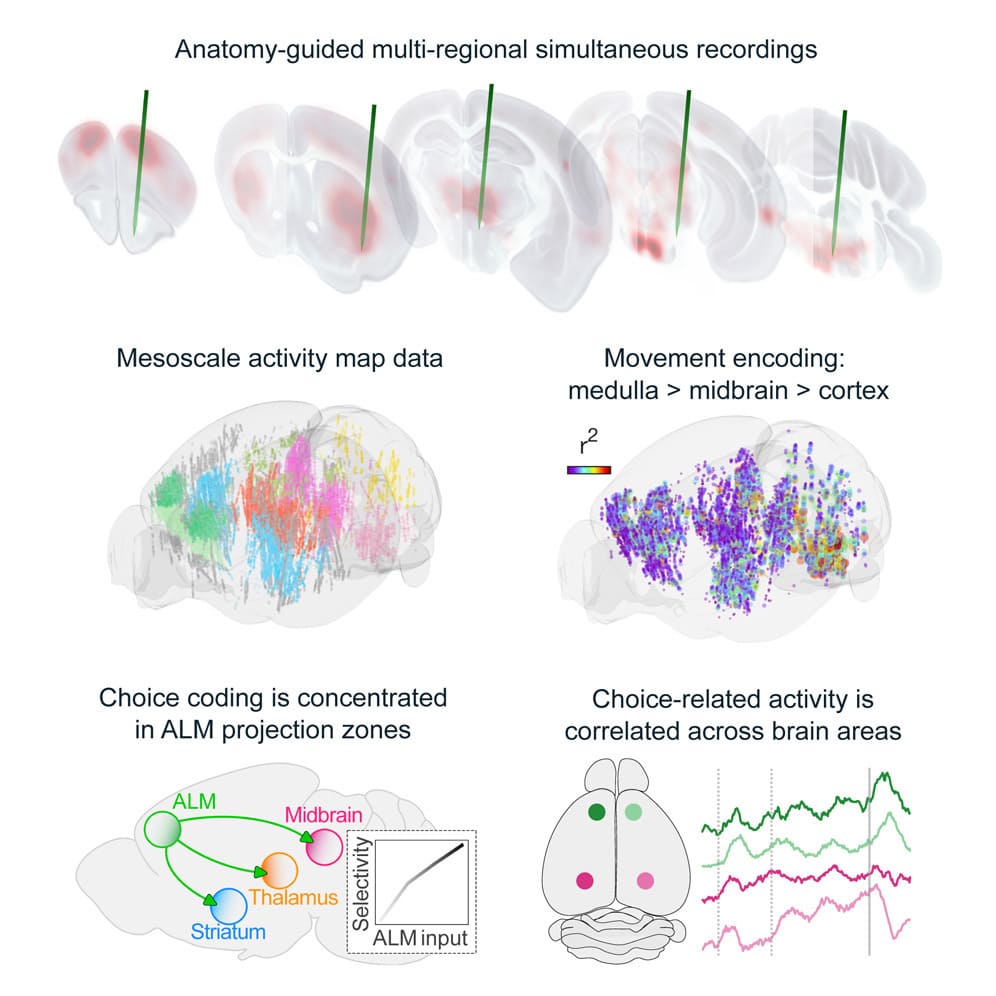
Behavior relies on activity in structured neural circuits that are distributed across the brain, but most experiments probe neurons in a single area at a time. Using multiple Neuropixels probes, we recorded from multi-regional loops connected to the anterior lateral motor cortex (ALM), a circuit node mediating memory-guided directional licking.
This dataset was collected for the Dendritic Coupling project, as part of the OpenScope project.
We developed a large-scale model of the multiregional mouse brain for a cardinal cognitive function called working memory, the brain’s ability to internally hold and process information without sensory input.

The OpenScope program publishes on an annual basis a detailed public report that summarizes all of our activities within the year. These reports highlight all developments of the year: new selected projects, completed projects, publications and technical developments of the platform.

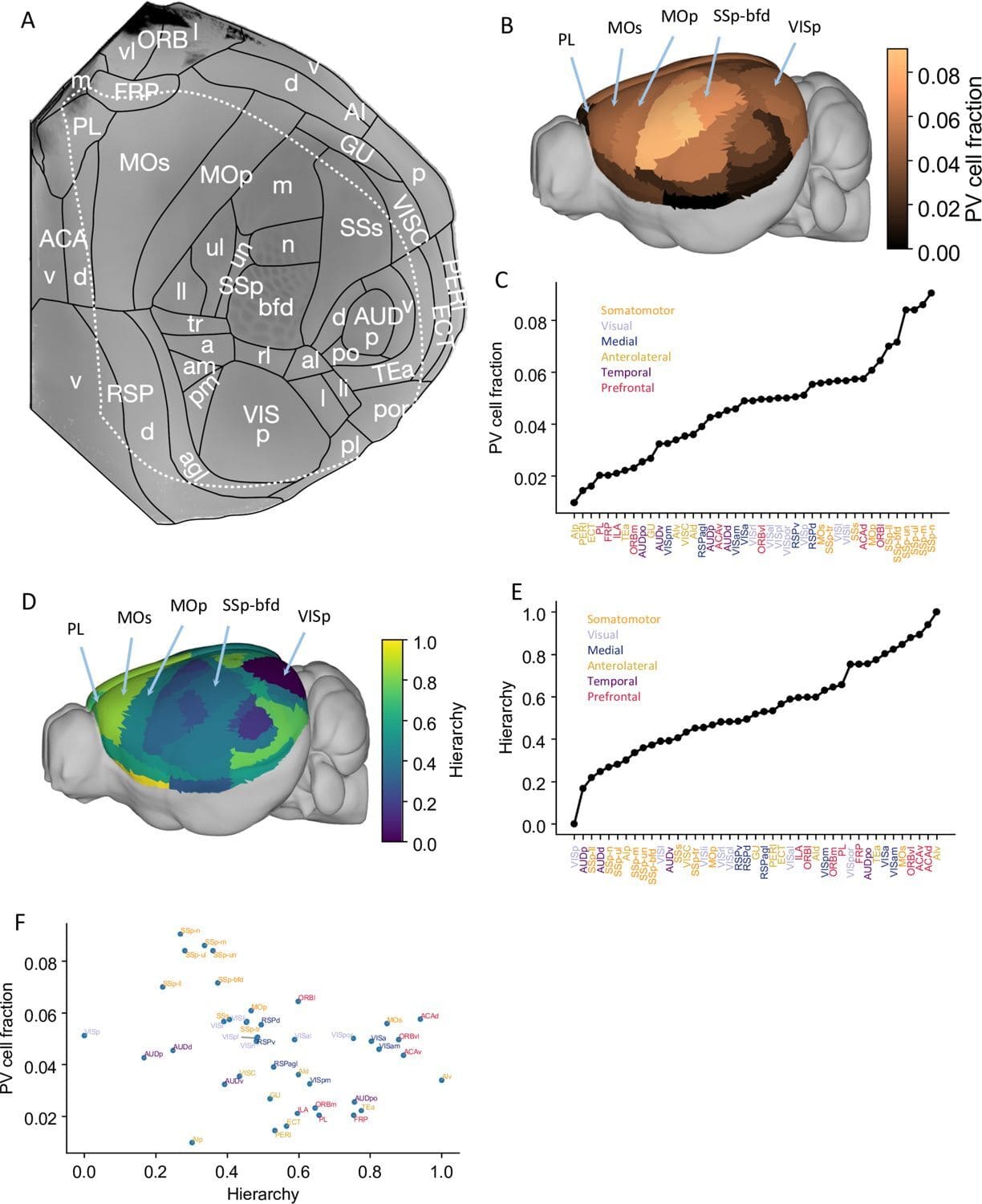
TIGRE2-jGCaMP8s-IRES-tTA2-WPRE knock-in mice co-express the jGCaMP8s genetically encoded calcium indicator (GECI) and a tetracycline controlled transactivator (tTA2) in a Cre recombinase-dependent manner from the intergenic Igs7 (TIGRE) locus.
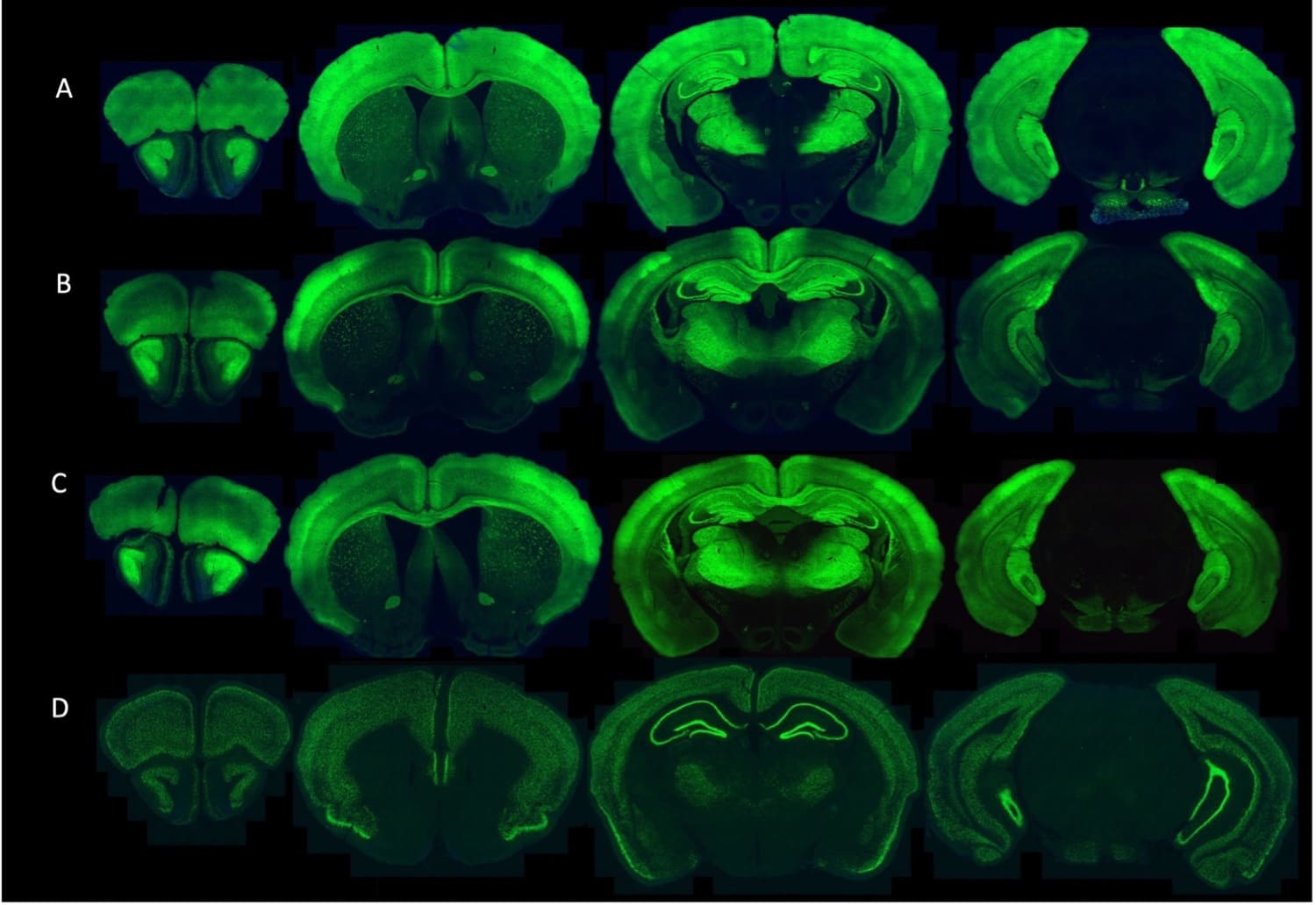
The GENIE Project Team at HHMI Janelia Research Campus have developed and characterized multiple transgenic mice expressing jGCaMP8s and jGCaMP8m.
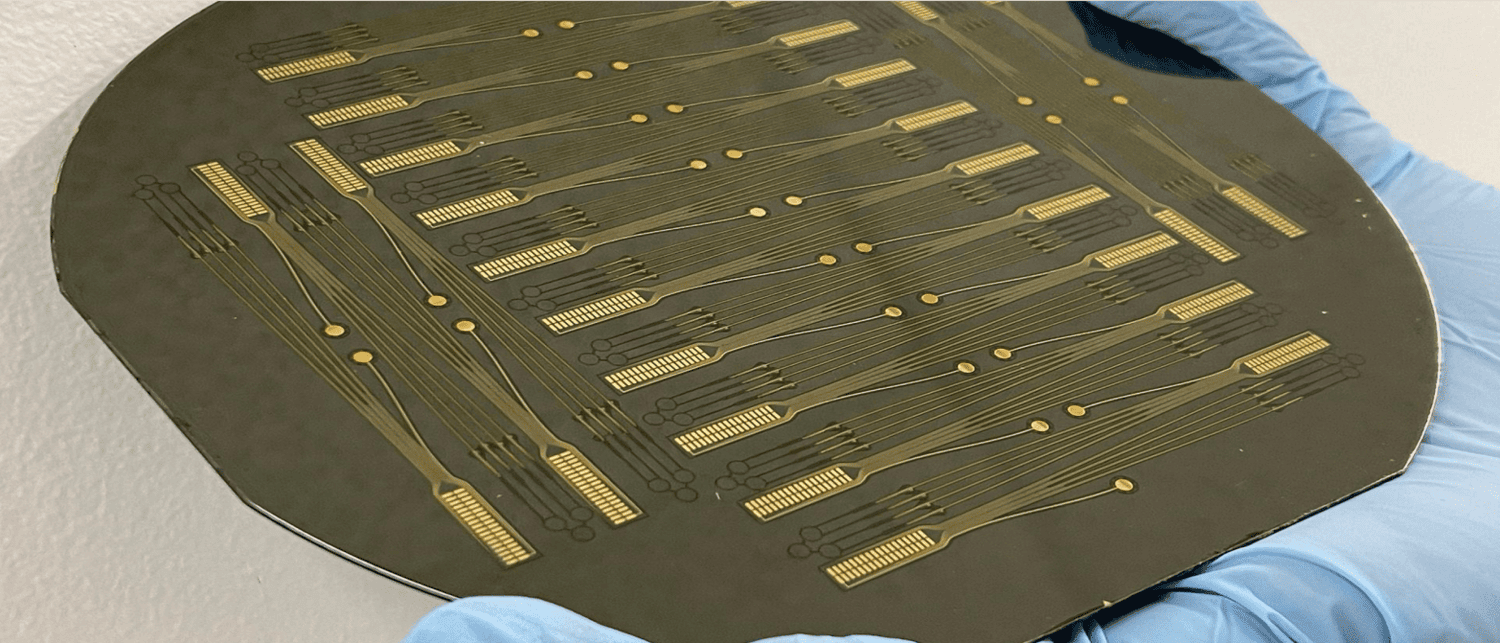
The Center for Advanced Motor BioEngineering and Research (CAMBER)'s mission is to create and disseminate next-generation tools for investigating motor control. They have created high-density electrode devices ("Myomatrix arrays”) that record muscle activity at high resolution across a wide range of muscles, species and behavior.
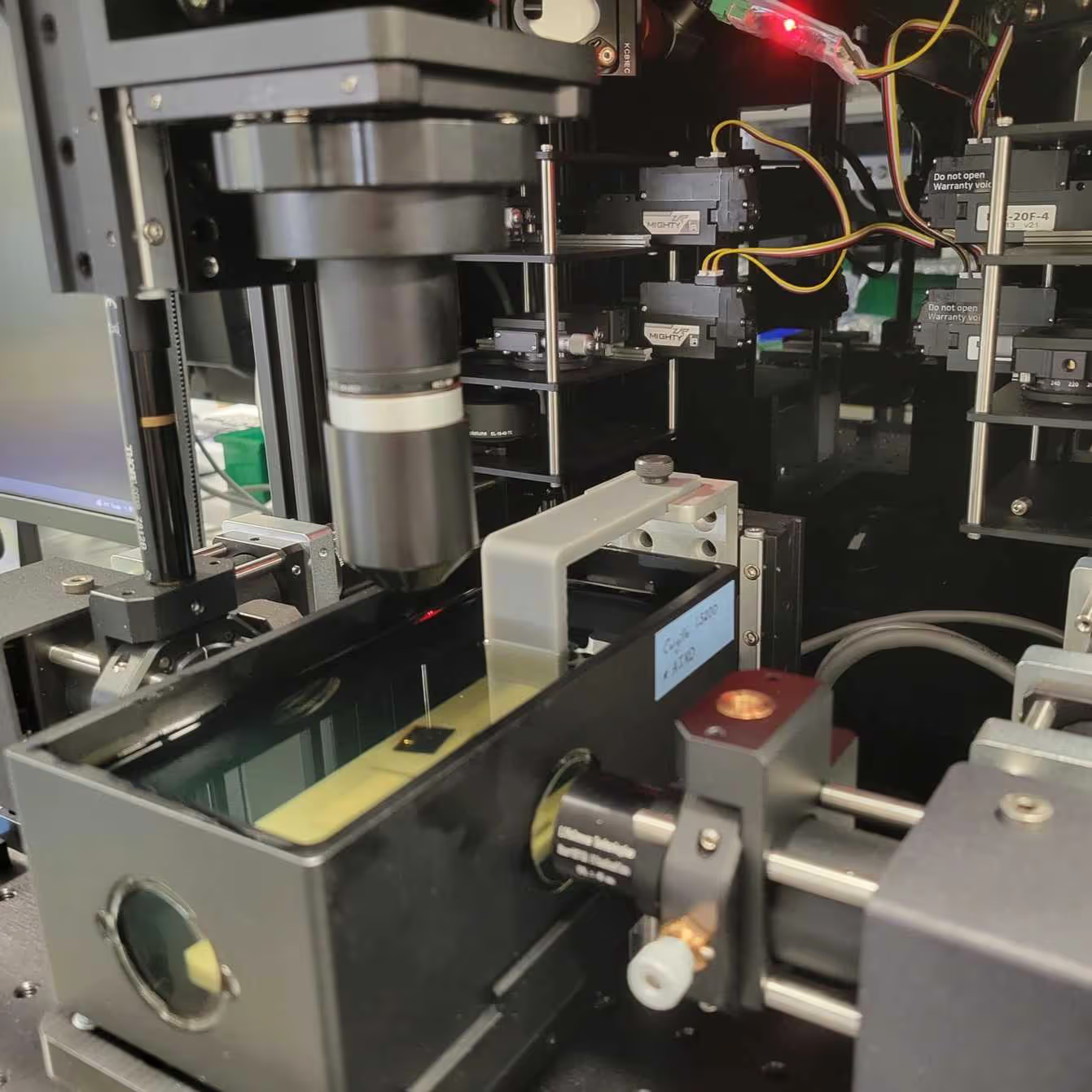
This protocol provides instructions to prepare the SmartSPIM microscope, to load samples, and to acquire data.

The SmartSPIM is an axially swept lightsheet microscope that produces uniform axial resolution across the entire imaging volume of large biological samples, such as whole mouse brains.
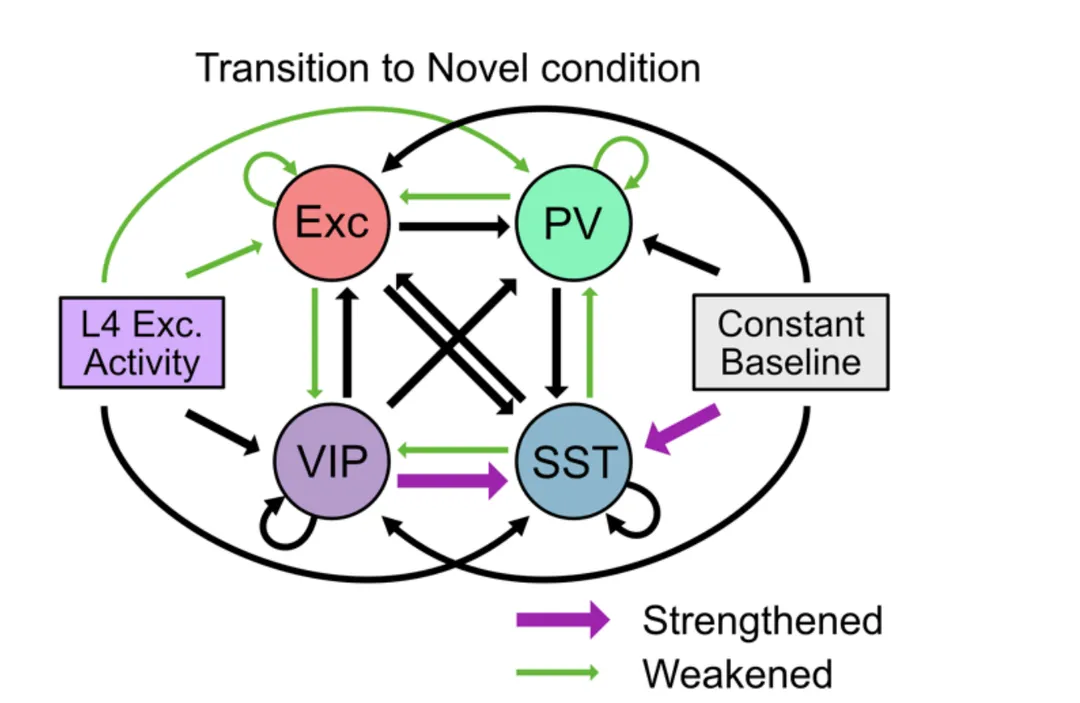
We employed a large-scale public 16 dataset of electrophysiological recordings in the visual cortex of awake, behaving mice using 17 Neuropixels probes and designed population network models to investigate the observed 18 changes in neural dynamics in response to a combination of distinct forms of novelty.
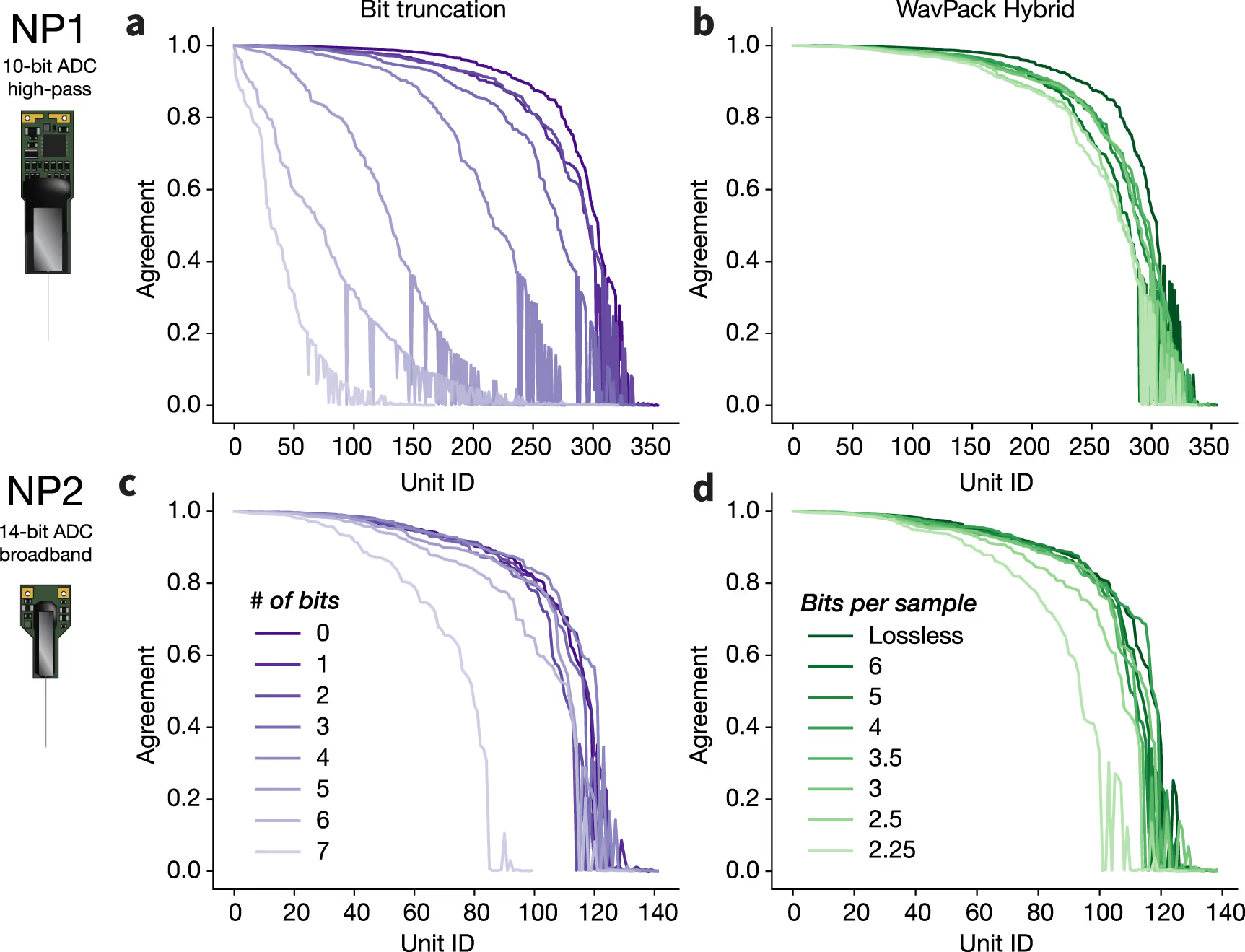
We establish a set of benchmarks for comparing the performance of various compression algorithms on experimental and simulated recordings from Neuropixels 1.0 (NP1) and 2.0 (NP2) probes.
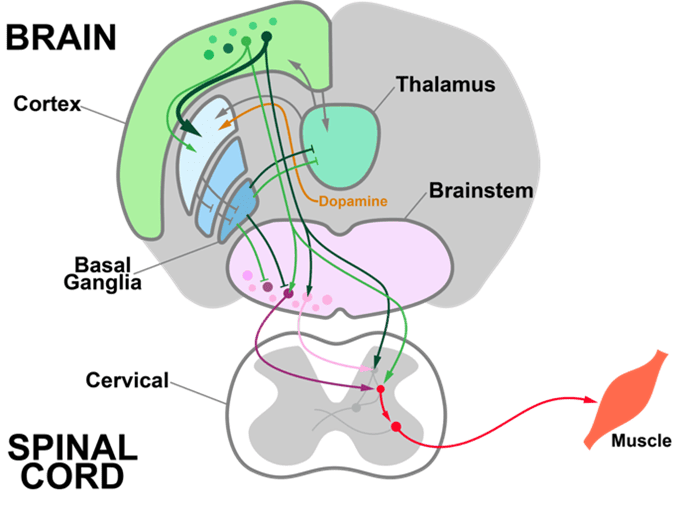
We are developing and using genetic, electrophysiological, optical, and behavioral approaches to investigate how the brain adaptively controls behavior. The team focuses on understanding the descending circuits that control the execution of actions and how they change when actions are reinforced and refined.

We are using large-scale electrophysiology to study how distributed brain regions coordinate their spiking activity to guide behavior in changing environments.

We are uncovering how neural signals are transmitted from subcortical brain regions via the thalamus to modulate cortical activity and thereby influence behavior.